Conjugated polymer nanoparticles for effective siRNA delivery to tobacco BY-2 protoplasts
- PMID: 21192827
- PMCID: PMC3023792
- DOI: 10.1186/1471-2229-10-291
Conjugated polymer nanoparticles for effective siRNA delivery to tobacco BY-2 protoplasts
Abstract
Background: Post transcriptional gene silencing (PTGS) is a mechanism harnessed by plant biologists to knock down gene expression. siRNAs contribute to PTGS that are synthesized from mRNAs or viral RNAs and function to guide cellular endoribonucleases to target mRNAs for degradation. Plant biologists have employed electroporation to deliver artificial siRNAs to plant protoplasts to study gene expression mechanisms at the single cell level. One drawback of electroporation is the extensive loss of viable protoplasts that occurs as a result of the transfection technology.
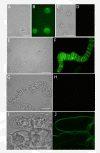
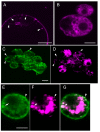
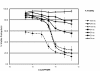
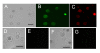
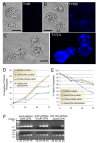
Results: We employed fluorescent conjugated polymer nanoparticles (CPNs) to deliver siRNAs and knockdown a target gene in plant protoplasts. CPNs are non toxic to protoplasts, having little impact on viability over a 72 h period. Microscopy and flow cytometry reveal that CPNs can penetrate protoplasts within 2 h of delivery. Cellular uptake of CPNs/siRNA complexes were easily monitored using epifluorescence microscopy. We also demonstrate that CPNs can deliver siRNAs targeting specific genes in the cellulose biosynthesis pathway (NtCesA-1a and NtCesA-1b).
Conclusions: While prior work showed that NtCesA-1 is a factor involved in cell wall synthesis in whole plants, we demonstrate that the same gene plays an essential role in cell wall regeneration in isolated protoplasts. Cell wall biosynthesis is central to cell elongation, plant growth and development. The experiments presented here shows that NtCesA is also a factor in cell viability. We show that CPNs are valuable vehicles for delivering siRNAs to plant protoplasts to study vital cellular pathways at the single cell level.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

