A pituitary-specific enhancer of the POMC gene with preferential activity in corticotrope cells
- PMID: 21193556
- PMCID: PMC5417307
- DOI: 10.1210/me.2010-0422
A pituitary-specific enhancer of the POMC gene with preferential activity in corticotrope cells
Abstract
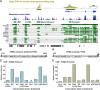
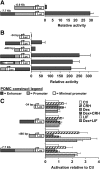
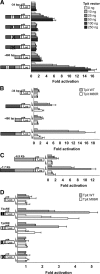
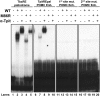
Cell-specific expression of the pituitary proopiomelanocortin (POMC) gene depends on the combination of tissue- and cell-restricted transcription factors such as Pitx1 and Tpit. These factors act on the proximal POMC promoter together with transcription factors that integrate inputs from signaling pathways. We now report the identification of an upstream enhancer in the POMC locus that is targeted by the same subset of transcription factors, except Pitx1. This enhancer located at -7 kb in the mouse POMC gene is highly dependent on Tpit for activity. Whereas Tpit requires Pitx1 for action on the promoter, it acts on the -7-kb enhancer as homodimers binding to a palindromic Tpit response element (TpitRE). Both half-sites of the TpitRE palindrome and Tpit homodimerization are required for activity. In vivo, the enhancer exhibits preferential activity in corticotrope cells of the anterior lobe whereas the promoter exhibits preference for intermediate lobe melanotropes. The enhancer is conserved among different species with the TpitRE palindrome localized at the center of conserved sequences. However, the mouse and human -7-kb enhancers do not exhibit conservation of hormone responsiveness and may differ in their relative importance for POMC expression. In summary, pituitary expression of the POMC gene relies on an upstream enhancer that complements the activity of the proximal promoter with Tpit as the major regulator of both regulatory regions.
Figures


References
-
- Drouin J. 2010. Pituitary development. In: , Melmed S (ed) The Pituitary, 3rd Edition. Elsevier-Academic Press, 3–19
-
- Tremblay JJ , Lanctôt C , Drouin J. 1998. The pan-pituitary activator of transcription, Ptx-1 (pituitary homeobox1), acts in synergy with SF-1 and Pit1 and is an upstream regulator of the Lim-homeodomain gene Lim3/Lhx3. Mol Endocrinol 12:428–441 - PubMed
-
- Sornson MW , Wu W , Dasen JS , Flynn SE , Norman DJ , O'Connell SM , Gukovsky I , Carriere C , Ryan AK , Miller AP , Zuo L , Gleiberman AS , Andersen B , Beamer WG , Rosenfeld MG. 1996. Pituitary lineage determination by the Prophet of Pit-1 homeodomain factor defective in Ames dwarfism. Nature 384:327–333 - PubMed
-
- Camper SA , Saunders TL , Katz RW , Reeves RH. 1990. The Pit-1 transcription factor gene is a candidate for the murine snell dwarf mutation. Genomics 8:586–590 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

