Assortment and packaging of the segmented rotavirus genome
- PMID: 21195621
- PMCID: PMC3072067
- DOI: 10.1016/j.tim.2010.12.002
Assortment and packaging of the segmented rotavirus genome
Abstract
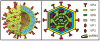
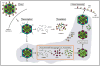
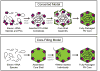
The rotavirus (RV) genome comprises 11 segments of double-stranded RNA (dsRNA) and is contained within a non-enveloped, icosahedral particle. During assembly, a highly coordinated selective packaging mechanism ensures that progeny RV virions contain one of each genome segment. Cis-acting signals thought to mediate assortment and packaging are associated with putative panhandle structures formed by base-pairing of the ends of RV plus-strand RNAs (+RNAs). Viral polymerases within assembling core particles convert the 11 distinct +RNAs to dsRNA genome segments. It remains unclear whether RV +RNAs are assorted before or during encapsidation, and the functions of viral proteins during these processes are not resolved. However, as reviewed here, recent insights gained from the study of RV and two other segmented RNA viruses, influenza A virus and bacteriophage Φ6, reveal potential mechanisms of RV assortment and packaging.
Published by Elsevier Ltd.
Figures

References
-
- Hundley F, et al. Genome rearrangements of bovine rotavirus after serial passage at high multiplicity of infection. Virology. 1985;143:88–103. - PubMed
-
- Joklik WK, Roner MR. What reassorts when reovirus genome segments reassort? J Bio Chem. 1995;270:4181–4184. - PubMed
-
- Patton JT. Evidence for equimolar synthesis of double-stranded RNA and minus- strand RNA in rotavirus-infected cells. Virus Res. 1990;17:199–208. - PubMed
-
- Hutchinson EC, et al. Genome packaging in influenza A virus. J Gen Virol. 2010;91:313–328. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

