A critical analysis of the combined usage of protein localization prediction methods: Increasing the number of independent data sets can reduce the accuracy of predicted mitochondrial localization
- PMID: 21195798
- PMCID: PMC3081538
- DOI: 10.1016/j.mito.2010.12.016
A critical analysis of the combined usage of protein localization prediction methods: Increasing the number of independent data sets can reduce the accuracy of predicted mitochondrial localization
Abstract
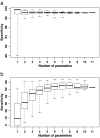
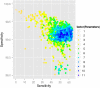
In the absence of a comprehensive experimentally derived mitochondrial proteome, several bioinformatic approaches have been developed to aid the identification of novel mitochondrial disease genes within mapped nuclear genetic loci. Often, many classifiers are combined to increase the sensitivity and specificity of the predictions. Here we show that the greatest sensitivity and specificity are obtained by using a combination of seven carefully selected classifiers. We also show that increasing the number of independent prediction methods can paradoxically decrease the accuracy of predicting mitochondrial localization. This approach will help to accelerate the identification of new mitochondrial disease genes by providing a principled way for the selection for combination of appropriate prediction methods of mitochondrial localization of proteins.
Copyright © 2011 Elsevier B.V. and Mitochondria Research Society. All rights reserved.
Figures


References
-
- Bannai H., Tamada Y., Maruyama O., Nakai K., Miyano S. Extensive feature detection of N-terminal protein sorting signals. Bioinformatics. 2002;18:298–305. - PubMed
-
- Baum P.A., Haussler D. What size net gives valid generalization? Neural Comput. 1989;1:150–160.
-
- Bourdon A., Minai L., Serre V., Jais J.P., Sarzi E., Aubert S., Chretien D., de Lonlay P., Paquis-Flucklinger V., Arakawa H., Nakamura Y., Munnich A., Rotig A. Mutation of RRM2B, encoding p53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nat. Genet. 2007;39:776–780. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

