Amyloid precursor protein (APP) processing genes and cerebrospinal fluid APP cleavage product levels in Alzheimer's disease
- PMID: 21196064
- PMCID: PMC3065534
- DOI: 10.1016/j.neurobiolaging.2010.10.020
Amyloid precursor protein (APP) processing genes and cerebrospinal fluid APP cleavage product levels in Alzheimer's disease
Abstract
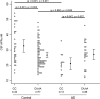
The aim of this exploratory investigation was to determine if genetic variation within amyloid precursor protein (APP) or its processing enzymes correlates with APP cleavage product levels: APPα, APPβ or Aβ42, in cerebrospinal fluid (CSF) of cognitively normal subjects or Alzheimer's disease (AD) patients. Cognitively normal control subjects (n = 170) and AD patients (n = 92) were genotyped for 19 putative regulatory tagging SNPs within 9 genes (APP, ADAM10, BACE1, BACE2, PSEN1, PSEN2, PEN2, NCSTN and APH1B) involved in the APP processing pathway. SNP genotypes were tested for their association with CSF APPα, APPβ, and Aβ42, AD risk and age-at-onset while taking into account age, gender, race and APOE ε4. After adjusting for multiple comparisons, a significant association was found between ADAM10 SNP rs514049 and APPα levels. In controls, the rs514049 CC genotype had higher APPα levels than the CA, AA collapsed genotype, whereas the opposite effect was seen in AD patients. These results suggest that genetic variation within ADAM10, an APP processing gene, influences CSF APPα levels in an AD specific manner.
Copyright © 2011 Elsevier Inc. All rights reserved.
Conflict of interest statement
Figures
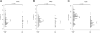
References
-
- Alves G, Bronnick K, Aarsland D, Blennow K, Zetterberg H, Ballard C, Kurz MW, Andreasson U, Tysnes OB, Larsen JP, Mulugeta E. CSF amyloid-beta and tau proteins, and cognitive performance, in early and untreated Parkinson’s disease: the Norwegian ParkWest study. J Neurol Neurosurg Psychiatry. 2010;81:1080–1086. - PubMed
-
- Baulac S, Lavoie MJ, Kimberly WT, Strahle J, Wolfe MS, Selkoe DJ, Xia W. Functional gamma-secretase complex assembly in Golgi/trans-Golgi network: interactions among presenilin, nicastrin, Aph1, Pen-2, and gamma-secretase substrates. Neurobiol Dis. 2003;14:194–204. - PubMed
-
- Bekris LM, Millard SP, Galloway NM, Vuletic S, Albers JJ, Li G, Galasko DR, Decarli C, Farlow MR, Clark CM, Quinn JF, Kaye JA, Schellenberg GD, Tsuang D, Peskind ER, Yu CE. Multiple SNPs within and surrounding the apolipoprotein E gene influence cerebrospinal fluid apolipoprotein E protein levels. J Alzheimers Dis. 2008;13:255–266. - PMC - PubMed
-
- Benito E, Barco A. CREB’s control of intrinsic and synaptic plasticity: implications for CREB-dependent memory models. Trends Neurosci. 2010;33:230–240. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

