Mechanism of chromosomal boundary action: roadblock, sink, or loop?
- PMID: 21196526
- PMCID: PMC3063668
- DOI: 10.1534/genetics.110.123752
Mechanism of chromosomal boundary action: roadblock, sink, or loop?
Abstract
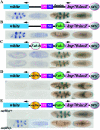
Boundary elements or insulators subdivide eukaryotic chromosomes into a series of structurally and functionally autonomous domains. They ensure that the action of enhancers and silencers is restricted to the domain in which these regulatory elements reside. Three models, the roadblock, sink/decoy, and topological loop, have been proposed to explain the insulating activity of boundary elements. Strong predictions about how boundaries will function in different experimental contexts can be drawn from these models. In the studies reported here, we have designed assays that test these predictions. The results of our assays are inconsistent with the expectations of the roadblock and sink models. Instead, they support the topological loop model.
© 2011 by the Genetics Society of America
Figures
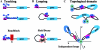
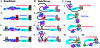





References
-
- Alfert, M., 1954. Composition and structure of giant chromosomes. Int. Rev. Cytol. 3 131–176.
-
- Barges, S., J. Mihaly, M. Galloni, K. Hagstrom, M. Muller et al., 2000. The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development 127 779–790. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

