Janus kinase activation by cytokine oncostatin M decreases PCSK9 expression in liver cells
- PMID: 21196532
- PMCID: PMC3035688
- DOI: 10.1194/jlr.M010603
Janus kinase activation by cytokine oncostatin M decreases PCSK9 expression in liver cells
Abstract
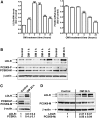
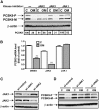
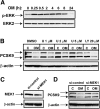
PCSK9 degrades LDL receptor (LDLR) in liver and thereby influences the circulating level of LDL cholesterol. Hence, mechanisms inhibiting PCSK9 expression have potential for cholesterol-lowering intervention. Previously, we demonstrated that oncostatin M (OM) activates LDLR gene transcription, resulting in an increased LDL uptake in HepG2 cells and a reduction of plasma LDL in hypercholesterolemic hamsters. Here we identify the suppression of PCSK9 expression by OM as one new mechanism that increases LDLR protein in HepG2 cells. Treating HepG2 cells with OM decreases PCSK9 mRNA and protein levels. Inhibition studies and small interfering RNA -targeted depletion revealed a critical role for JAK1 and JAK2 in mediating this OM inhibitory effect. Furthermore, we showed that OM induces transient phosphorylation of STAT1, STAT3, and STAT5 and sustained activation of ERK signaling molecules. While depletion of STAT members in HepG2 cells did not affect OM inhibitory activity on PCSK9 expression, blocking activation of the MEK1/ERK signaling pathway resulted in attenuation of the OM inhibitory effect. Finally, by using an anti-hamster PCSK9 antibody, we demonstrated the in vivo suppression of liver PCSK9 mRNA and protein expression by OM in hypercholesterolemic hamsters. Our study uncovered a cytokine-triggered regulatory network for PCSK9 expression that is linked to JAKs and the ERK signaling pathway.
Figures





References
-
- Seidah N. G., Benjannet S., Wickham L., Marcinkiewicz J., Bélanger J., Stifani S., Basak A., Prat A., Chrétien M. 2003. The secretory proprotein convertase neutal apoptosis-regulated convertase 1 (NARC-1): liver regeneration and neuronal differentiation. Proc. Natl. Acad. Sci. U S A. 100: 928–933. - PMC - PubMed
-
- Abifadel M., Varret M., Rabes J. P., Allard D., Ouguerram K., Devillers M., Cruaud C., Benjannet S., Wickham L., Erlich D., et al. 2003. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat. Genet. 34: 154–156. - PubMed
-
- Maxwell K. N., Soccio R. E., Duncan E. M., Sehayek E., Breslow J. L. 2003. Novel putative SREBP and LXR target genes identified by microarray analysis in liver of cholesterol-fed mice. J. Lipid Res. 44: 2109–2119. - PubMed
-
- Benjannet S., Rhainds D., Essalmani R., Mayne J., Wickham L., Jin W., Asselin M. C., Hamelin J., Varret M., Allard D., et al. 2004. NARC-1/PCSK9 and its natural mutants: zymogen cleavage and effects on the low density lipoprotein (LDL) receptor and LDL cholesterol. J. Biol. Chem. 279: 48865–48875. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

