MTML-msBayes: approximate Bayesian comparative phylogeographic inference from multiple taxa and multiple loci with rate heterogeneity
- PMID: 21199577
- PMCID: PMC3031198
- DOI: 10.1186/1471-2105-12-1
MTML-msBayes: approximate Bayesian comparative phylogeographic inference from multiple taxa and multiple loci with rate heterogeneity
Abstract
Background: MTML-msBayes uses hierarchical approximate Bayesian computation (HABC) under a coalescent model to infer temporal patterns of divergence and gene flow across codistributed taxon-pairs. Under a model of multiple codistributed taxa that diverge into taxon-pairs with subsequent gene flow or isolation, one can estimate hyper-parameters that quantify the mean and variability in divergence times or test models of migration and isolation. The software uses multi-locus DNA sequence data collected from multiple taxon-pairs and allows variation across taxa in demographic parameters as well as heterogeneity in DNA mutation rates across loci. The method also allows a flexible sampling scheme: different numbers of loci of varying length can be sampled from different taxon-pairs.
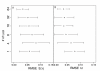
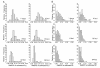
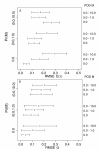
Results: Simulation tests reveal increasing power with increasing numbers of loci when attempting to distinguish temporal congruence from incongruence in divergence times across taxon-pairs. These results are robust to DNA mutation rate heterogeneity. Estimating mean divergence times and testing simultaneous divergence was less accurate with migration, but improved if one specified the correct migration model. Simulation validation tests demonstrated that one can detect the correct migration or isolation model with high probability, and that this HABC model testing procedure was greatly improved by incorporating a summary statistic originally developed for this task (Wakeley's ΨW). The method is applied to an empirical data set of three Australian avian taxon-pairs and a result of simultaneous divergence with some subsequent gene flow is inferred.
Conclusions: To retain flexibility and compatibility with existing bioinformatics tools, MTML-msBayes is a pipeline software package consisting of Perl, C and R programs that are executed via the command line. Source code and binaries are available for download at http://msbayes.sourceforge.net/ under an open source license (GNU Public License).
Figures
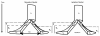



References
-
- Bermingham E, Moritz C. Comparative phylogeography: concepts and applications. Mol Ecol. 1998;7:367–369. doi: 10.1046/j.1365-294x.1998.00424.x. - DOI
-
- Arbogast BS, Kenagy GJ. Comparative phylogeography as an integrative approach to historical biogeography. J Biogeogr. 2001;28:819–825. doi: 10.1046/j.1365-2699.2001.00594.x. - DOI
-
- Coyne JA, Orr HA. Speciation. Sunderland, MA: Sinauer Associates Inc; 2004.
-
- Avise JC. Phylogeography: The history and formation of species. Cambridge: Harvard University Press; 2000.
-
- Hubbell SP. The Unified Neutral Theory of Biodiversity and Biogeography. Princeton, NJ: Princeton University Press; 2001.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

