Polymorphisms in an intronic region of the myocilin gene associated with primary open-angle glaucoma--a possible role for alternate splicing
- PMID: 21203411
- PMCID: PMC3013065
Polymorphisms in an intronic region of the myocilin gene associated with primary open-angle glaucoma--a possible role for alternate splicing
Abstract
Purpose: To examine the possible role of alternate splicing leading to aggregation of myocilin in primary open-angle glaucoma.
Methods: Several single nucleotide variations found in the myocilin (MYOC) genomic region were collected and examined for their possible role in causing splice-site alterations. A model for myocilin built using a knowledge-based consensus method was used to map the altered protein products. A total of 150 open-angle glaucoma patients and 50 normal age-matched control subjects were screened for the predicted polymorphisms, and clustering was performed.
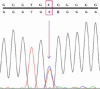
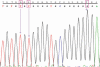
Results: A total of 124 genomic variations were screened, and six polymorphisms that lead to altered protein products were detected as possible candidates for the alternative splicing mechanism. Five of these lay in the intronic regions, and the one that lay in the exon region corresponded to the previously identified polymorphism (Tyr347Tyr) implicated in primary open-angle glaucoma. Experimentally screening the intronic region of the MYOC gene showed the presence of the predicted g.14072G>A polymorphism, g.1293C/T heterozygous polymorphism, instead of our predicted g.1293C/- polymorphism. Other than the prediction, two novel SNPs (g.1295G>T and g.1299T>G) and two reported SNPs (g.1284G>T and g.1286G>T) were also identified. Cluster analysis showed the g.14072G>A homozygous condition was more common in this cohort than the heterozygous condition.
Conclusions: We previously proposed that the disruption of dimer or oligomer formation by the C-term region allows greater chances of nucleation for aggregation. Here we suggest that polymorphisms in the myocilin genomic region that cause synonymous codon changes or those that occur in the intron regions can possibly lead to altered myocilin protein products through altered intron-exon splicing.
Figures



Similar articles
-
Evaluation and understanding of myocilin mutations in Indian primary open angle glaucoma patients.Mol Vis. 2003 Nov 14;9:606-14. Mol Vis. 2003. PMID: 14627955
-
Mutations in MYOC gene of Indian primary open angle glaucoma patients.Mol Vis. 2002 Nov 15;8:442-8. Mol Vis. 2002. PMID: 12447164
-
Identification of mutations in the myocilin (MYOC) gene in Taiwanese patients with juvenile-onset open-angle glaucoma.Mol Vis. 2007 Sep 10;13:1627-34. Mol Vis. 2007. PMID: 17893664
-
Current perspectives on the TIGR/MYOC gene (Myocilin) and glaucoma.Ophthalmol Clin North Am. 2003 Dec;16(4):515-27, v-vi. doi: 10.1016/s0896-1549(03)00068-3. Ophthalmol Clin North Am. 2003. PMID: 14740993 Review.
-
Genetic dissection of myocilin glaucoma.Hum Mol Genet. 2004 Apr 1;13 Spec No 1:R91-102. doi: 10.1093/hmg/ddh074. Epub 2004 Feb 5. Hum Mol Genet. 2004. PMID: 14764620 Review.
Cited by
-
Glial cell line-derived neurotrophic factor (GDNF) as a novel candidate gene of anxiety.PLoS One. 2013 Dec 6;8(12):e80613. doi: 10.1371/journal.pone.0080613. eCollection 2013. PLoS One. 2013. PMID: 24324616 Free PMC article.
-
The mutational spectrum of Myocilin gene among familial versus sporadic cases of Juvenile onset open angle glaucoma.Eye (Lond). 2021 Feb;35(2):400-408. doi: 10.1038/s41433-020-0850-z. Epub 2020 Apr 16. Eye (Lond). 2021. PMID: 32300215 Free PMC article.
-
Comprehensive sequencing of the myocilin gene in a selected cohort of severe primary open-angle glaucoma patients.Sci Rep. 2019 Feb 28;9(1):3100. doi: 10.1038/s41598-019-38760-y. Sci Rep. 2019. PMID: 30816137 Free PMC article.
References
-
- Goldwyn R, Waltman SR, Becker B. Primary open-angle glaucoma in adolescents and young adults. Arch Ophthalmol. 1970;84:579–82. - PubMed
-
- Johnson AT, Drack AV, Kwitek AE, Cannon RL, Stone EM, Alward WL. Clinical features and linkage analysis of a family with autosomal dominant juvenile glaucoma. Ophthalmology. 1993;100:524–9. - PubMed
-
- Quigley HA. Open-angle glaucoma. N Engl J Med. 1993;328:1097–106. - PubMed
-
- Quigley HA. Proportion of those with open-angle glaucoma who become blind. Ophthalmology. 1999;106:2039–41. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
