Structure-function relationship of cytoplasmic and nuclear IκB proteins: an in silico analysis
- PMID: 21203422
- PMCID: PMC3009747
- DOI: 10.1371/journal.pone.0015782
Structure-function relationship of cytoplasmic and nuclear IκB proteins: an in silico analysis
Abstract
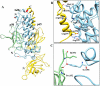
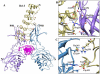
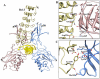
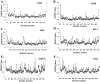
Cytoplasmic IκB proteins are primary regulators that interact with NF-κB subunits in the cytoplasm of unstimulated cells. Upon stimulation, these IκB proteins are rapidly degraded, thus allowing NF-κB to translocate into the nucleus and activate the transcription of genes encoding various immune mediators. Subsequent to translocation, nuclear IκB proteins play an important role in the regulation of NF-κB transcriptional activity by acting either as activators or inhibitors. To date, molecular basis for the binding of IκBα, IκBβ and IκBζ along with their partners is known; however, the activation and inhibition mechanism of the remaining IκB (IκBNS, IκBε and Bcl-3) proteins remains elusive. Moreover, even though IκB proteins are structurally similar, it is difficult to determine the exact specificities of IκB proteins towards their respective binding partners. The three-dimensional structures of IκBNS, IκBζ and IκBε were modeled. Subsequently, we used an explicit solvent method to perform detailed molecular dynamic simulations of these proteins along with their known crystal structures (IκBα, IκBβ and Bcl-3) in order to investigate the flexibility of the ankyrin repeat domains (ARDs). Furthermore, the refined models of IκBNS, IκBε and Bcl-3 were used for multiple protein-protein docking studies for the identification of IκBNS-p50/p50, IκBε-p50/p65 and Bcl-3-p50/p50 complexes in order to study the structural basis of their activation and inhibition. The docking experiments revealed that IκBε masked the nuclear localization signal (NLS) of the p50/p65 subunits, thereby preventing its translocation into the nucleus. For the Bcl-3- and IκBNS-p50/p50 complexes, the results show that Bcl-3 mediated transcription through its transactivation domain (TAD) while IκBNS inhibited transcription due to its lack of a TAD, which is consistent with biochemical studies. Additionally, the numbers of identified flexible residues were equal in number among all IκB proteins, although they were not conserved. This could be the primary reason for their binding partner specificities.
Conflict of interest statement
Figures





References
-
- Hoffmann A, Baltimore D. Circuitry of nuclear factor kappaB signaling. Immunol Rev. 2006;210:171–186. - PubMed
-
- Ghosh S, Hayden MS. New regulators of NF-kappaB in inflammation. Nat Rev Immunol. 2008;8:837–848. - PubMed
-
- Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. - PubMed
-
- Yamamoto M, Takeda K. Role of nuclear IkappaB proteins in the regulation of host immune responses. J Infect Chemother. 2008;14:265–269. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

