ATL9, a RING zinc finger protein with E3 ubiquitin ligase activity implicated in chitin- and NADPH oxidase-mediated defense responses
- PMID: 21203445
- PMCID: PMC3009710
- DOI: 10.1371/journal.pone.0014426
ATL9, a RING zinc finger protein with E3 ubiquitin ligase activity implicated in chitin- and NADPH oxidase-mediated defense responses
Abstract
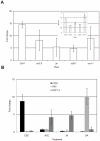
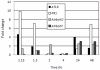
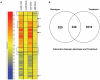
Pathogen associated molecular patterns (PAMPs) are signals detected by plants that activate basal defenses. One of these PAMPs is chitin, a carbohydrate present in the cell walls of fungi and in insect exoskeletons. Previous work has shown that chitin treatment of Arabidopsis thaliana induced defense-related genes in the absence of a pathogen and that the response was independent of the salicylic acid (SA), jasmonic acid (JA) and ethylene (ET) signaling pathways. One of these genes is ATL9 ( = ATL2G), which encodes a RING zinc-finger like protein. In the current work we demonstrate that ATL9 has E3 ubiquitin ligase activity and is localized to the endoplasmic reticulum. The expression pattern of ATL9 is positively correlated with basal defense responses against Golovinomyces cichoracearum, a biotrophic fungal pathogen. The basal levels of expression and the induction of ATL9 by chitin, in wild type plants, depends on the activity of NADPH oxidases suggesting that chitin-mediated defense response is NADPH oxidase dependent. Although ATL9 expression is not induced by treatment with known defense hormones (SA, JA or ET), full expression in response to chitin is compromised slightly in mutants where ET- or SA-dependent signaling is suppressed. Microarray analysis of the atl9 mutant revealed candidate genes that appear to act downstream of ATL9 in chitin-mediated defenses. These results hint at the complexity of chitin-mediated signaling and the potential interplay between elicitor-mediated signaling, signaling via known defense pathways and the oxidative burst.
Conflict of interest statement
Figures






References
-
- Flor HH. Current status of the gene-for-gene concept. Annu Rev Phytopathol. 1971;9:275–296.
-
- Gomez-Gomez L. Plant perception systems for pathogen recognition and defence. Molecular Immunology. 2004;41:1055–1062. - PubMed
-
- Libault M, Wan JR, Czechowski T, Udvardi M, Stacey G. Identification of 118 Arabidopsis transcription factor and 30 ubiquitin-ligase genes responding to chitin, a plant-defense elicitor. Molecular Plant-Microbe Interactions. 2007;20:900–911. - PubMed

