Molecular architectures of trimeric SIV and HIV-1 envelope glycoproteins on intact viruses: strain-dependent variation in quaternary structure
- PMID: 21203482
- PMCID: PMC3009598
- DOI: 10.1371/journal.ppat.1001249
Molecular architectures of trimeric SIV and HIV-1 envelope glycoproteins on intact viruses: strain-dependent variation in quaternary structure
Abstract
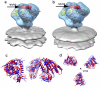
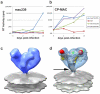
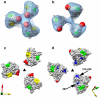
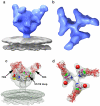
The initial step in target cell infection by human, and the closely related simian immunodeficiency viruses (HIV and SIV, respectively) occurs with the binding of trimeric envelope glycoproteins (Env), composed of heterodimers of the viral transmembrane glycoprotein (gp41) and surface glycoprotein (gp120) to target T-cells. Knowledge of the molecular structure of trimeric Env on intact viruses is important both for understanding the molecular mechanisms underlying virus-cell interactions and for the design of effective immunogen-based vaccines to combat HIV/AIDS. Previous analyses of intact HIV-1 BaL virions have already resulted in structures of trimeric Env in unliganded and CD4-liganded states at ~20 Å resolution. Here, we show that the molecular architectures of trimeric Env from SIVmneE11S, SIVmac239 and HIV-1 R3A strains are closely comparable to that previously determined for HIV-1 BaL, with the V1 and V2 variable loops located at the apex of the spike, close to the contact zone between virus and cell. The location of the V1/V2 loops in trimeric Env was definitively confirmed by structural analysis of HIV-1 R3A virions engineered to express Env with deletion of these loops. Strikingly, in SIV CP-MAC, a CD4-independent strain, trimeric Env is in a constitutively "open" conformation with gp120 trimers splayed out in a conformation similar to that seen for HIV-1 BaL Env when it is complexed with sCD4 and the CD4i antibody 17b. Our findings suggest a structural explanation for the molecular mechanism of CD4-independent viral entry and further establish that cryo-electron tomography can be used to discover distinct, functionally relevant quaternary structures of Env displayed on intact viruses.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. - PubMed
-
- Dalgleish AG, Beverley PC, Clapham PR, Crawford DH, Greaves MF, et al. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312:763–767. - PubMed
-
- Burton DR, Desrosiers RC, Doms RW, Koff WC, Kwong PD, et al. HIV vaccine design and the neutralizing antibody problem. Nat Immunol. 2004;5:233–236. - PubMed
-
- Sato S, Johnson W. Antibody-mediated neutralization and Simian immunodeficiency virus models of HIV/AIDS. Current HIV Research. 2007;5:594–607. - PubMed
-
- Chen B, Vogan EM, Gong H, Skehel JJ, Wiley DC, et al. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature. 2005;433:834–841. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

