Phosphorylation mechanism and structure of serine-arginine protein kinases
- PMID: 21205204
- PMCID: PMC3079193
- DOI: 10.1111/j.1742-4658.2010.07992.x
Phosphorylation mechanism and structure of serine-arginine protein kinases
Abstract
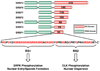
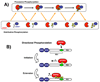
The splicing of mRNA requires a group of essential factors known as SR proteins, which participate in the maturation of the spliceosome. These proteins contain one or two RNA recognition motifs and a C-terminal domain rich in Arg-Ser repeats (RS domain). SR proteins are phosphorylated at numerous serines in the RS domain by the SR-specific protein kinase (SRPK) family of protein kinases. RS domain phosphorylation is necessary for entry of SR proteins into the nucleus, and may also play important roles in alternative splicing, mRNA export, and other processing events. Although SR proteins are polyphosphorylated in vivo, the mechanism underlying this complex reaction has only been recently elucidated. Human alternative splicing factor [serine/arginine-rich splicing factor 1 (SRSF1)], a prototype for the SR protein family, is regiospecifically phosphorylated by SRPK1, a post-translational modification that controls cytoplasmic-nuclear localization. SRPK1 binds SRSF1 with unusually high affinity, and rapidly modifies about 10-12 serines in the N-terminal region of the RS domain (RS1), using a mechanism that incorporates sequential, C-terminal to N-terminal phosphorylation and several processive steps. SRPK1 employs a highly dynamic feeding mechanism for RS domain phosphorylation in which the N-terminal portion of RS1 is initially bound to a docking groove in the large lobe of the kinase domain. Upon subsequent rounds of phosphorylation, this N-terminal segment translocates into the active site, and a β-strand in RNA recognition motif 2 unfolds and occupies the docking groove. These studies indicate that efficient regiospecific phosphorylation of SRSF1 is the result of a contoured binding cavity in SRPK1, a lengthy Arg-Ser repetitive segment in the RS domain, and a highly directional processing mechanism.
Journal compilation © 2011 FEBS. No claim to original US government works.
Figures



References
-
- Venables JP. Unbalanced alternative splicing and its significance in cancer. Bioessays. 2006;28:378–386. - PubMed
-
- Faustino NA, Cooper TA. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. - PubMed
-
- Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell. 2003;12:5–14. - PubMed
-
- Wu JY, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

