C. elegans bicd-1, homolog of the Drosophila dynein accessory factor Bicaudal D, regulates the branching of PVD sensory neuron dendrites
- PMID: 21205795
- PMCID: PMC3014636
- DOI: 10.1242/dev.060939
C. elegans bicd-1, homolog of the Drosophila dynein accessory factor Bicaudal D, regulates the branching of PVD sensory neuron dendrites
Abstract
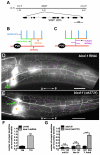
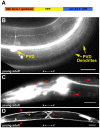
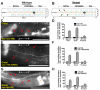
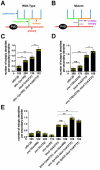
The establishment of cell type-specific dendritic arborization patterns is a key phase in the assembly of neuronal circuitry that facilitates the integration and processing of synaptic and sensory input. Although studies in Drosophila and vertebrate systems have identified a variety of factors that regulate dendrite branch formation, the molecular mechanisms that control this process remain poorly defined. Here, we introduce the use of the Caenorhabditis elegans PVD neurons, a pair of putative nociceptors that elaborate complex dendritic arbors, as a tractable model for conducting high-throughput RNAi screens aimed at identifying key regulators of dendritic branch formation. By carrying out two separate RNAi screens, a small-scale candidate-based screen and a large-scale screen of the ~3000 genes on chromosome IV, we retrieved 11 genes that either promote or suppress the formation of PVD-associated dendrites. We present a detailed functional characterization of one of the genes, bicd-1, which encodes a microtubule-associated protein previously shown to modulate the transport of mRNAs and organelles in a variety of organisms. Specifically, we describe a novel role for bicd-1 in regulating dendrite branch formation and show that bicd-1 is likely to be expressed, and primarily required, in PVD neurons to control dendritic branching. We also present evidence that bicd-1 operates in a conserved pathway with dhc-1 and unc-116, components of the dynein minus-end-directed and kinesin-1 plus-end-directed microtubule-based motor complexes, respectively, and interacts genetically with the repulsive guidance receptor unc-5.
Figures




References
-
- Ashrafi K., Chang F. Y., Watts J. L., Fraser A. G., Kamath R. S., Ahringer J., Ruvkun G. (2003). Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature 421, 268-272 - PubMed
-
- Baens M., Marynen P. (1997). A human homologue (BICD1) of the Drosophila bicaudal-D gene. Genomics 45, 601-606 - PubMed
-
- Bullock S. L., Ish-Horowicz D. (2001). Conserved signals and machinery for RNA transport in Drosophila oogenesis and embryogenesis. Nature 414, 611-616 - PubMed
-
- Bullock S. L., Nicol A., Gross S. P., Zicha D. (2006). Guidance of bidirectional motor complexes by mRNA cargoes through control of dynein number and activity. Curr. Biol. 16, 1447-1452 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

