Gene inactivation and its implications for annotation in the era of personal genomics
- PMID: 21205862
- PMCID: PMC3012931
- DOI: 10.1101/gad.1968411
Gene inactivation and its implications for annotation in the era of personal genomics
Abstract
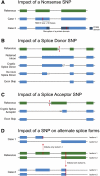
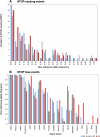
The first wave of personal genomes documents how no single individual genome contains the full complement of functional genes. Here, we describe the extent of variation in gene and pseudogene numbers between individuals arising from inactivation events such as premature termination or aberrant splicing due to single-nucleotide polymorphisms. This highlights the inadequacy of the current reference sequence and gene set. We present a proposal to define a reference gene set that will remain stable as more individuals are sequenced. In particular, we recommend that the ancestral allele be used to define the reference sequence from which a core human reference gene annotation set can be derived. In addition, we call for the development of an expanded gene set to include human-specific genes that have arisen recently and are absent from the ancestral set.
Figures



References
-
- Aoshima M, Nunoi H, Shimazu M, Shimizu S, Tatsuzawa O, Kenney RT, Kanegasaki S 1996. Two-exon skipping due to a point mutation in p67-phox–deficient chronic granulomatous disease. Blood 88: 1841–1845 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
