Observation and prediction of recurrent human translocations mediated by NAHR between nonhomologous chromosomes
- PMID: 21205869
- PMCID: PMC3012924
- DOI: 10.1101/gr.111609.110
Observation and prediction of recurrent human translocations mediated by NAHR between nonhomologous chromosomes
Abstract
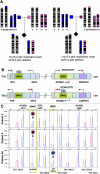
Four unrelated families with the same unbalanced translocation der(4)t(4;11)(p16.2;p15.4) were analyzed. Both of the breakpoint regions in 4p16.2 and 11p15.4 were narrowed to large ∼359-kb and ∼215-kb low-copy repeat (LCR) clusters, respectively, by aCGH and SNP array analyses. DNA sequencing enabled mapping the breakpoints of one translocation to 24 bp within interchromosomal paralogous LCRs of ∼130 kb in length and 94.7% DNA sequence identity located in olfactory receptor gene clusters, indicating nonallelic homologous recombination (NAHR) as the mechanism for translocation formation. To investigate the potential involvement of interchromosomal LCRs in recurrent chromosomal translocation formation, we performed computational genome-wide analyses and identified 1143 interchromosomal LCR substrate pairs, >5 kb in size and sharing >94% sequence identity that can potentially mediate chromosomal translocations. Additional evidence for interchromosomal NAHR mediated translocation formation was provided by sequencing the breakpoints of another recurrent translocation, der(8)t(8;12)(p23.1;p13.31). The NAHR sites were mapped within 55 bp in ∼7.8-kb paralogous subunits of 95.3% sequence identity located in the ∼579-kb (chr 8) and ∼287-kb (chr 12) LCR clusters. We demonstrate that NAHR mediates recurrent constitutional translocations t(4;11) and t(8;12) and potentially many other interchromosomal translocations throughout the human genome. Furthermore, we provide a computationally determined genome-wide "recurrent translocation map."
Figures





References
-
- Abeysinghe SS, Chuzhanova N, Krawczak M, Ball EV, Cooper DN 2003. Translocation and gross deletion breakpoints in human inherited disease and cancer I: Nucleotide composition and recombination-associated motifs. Hum Mutat 22: 229–244 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
