Tumour gene expression predicts response to cetuximab in patients with KRAS wild-type metastatic colorectal cancer
- PMID: 21206494
- PMCID: PMC3049558
- DOI: 10.1038/sj.bjc.6606054
Tumour gene expression predicts response to cetuximab in patients with KRAS wild-type metastatic colorectal cancer
Abstract
Background: Although it is accepted that metastatic colorectal cancers (mCRCs) that carry activating mutations in KRAS are unresponsive to anti-epidermal growth factor receptor (EGFR) monoclonal antibodies, a significant fraction of KRAS wild-type (wt) mCRCs are also unresponsive to anti-EGFR therapy. Genes encoding EGFR ligands amphiregulin (AREG) and epiregulin (EREG) are promising gene expression-based markers but have not been incorporated into a test to dichotomise KRAS wt mCRC patients with respect to sensitivity to anti-EGFR treatment.
Methods: We used RT-PCR to test 110 candidate gene expression markers in primary tumours from 144 KRAS wt mCRC patients who received monotherapy with the anti-EGFR antibody cetuximab. Results were correlated with multiple clinical endpoints: disease control, objective response, and progression-free survival (PFS).
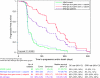
Results: Expression of many of the tested candidate genes, including EREG and AREG, strongly associate with all clinical endpoints. Using multivariate analysis with two-layer five-fold cross-validation, we constructed a four-gene predictive classifier. Strikingly, patients below the classifier cutpoint had PFS and disease control rates similar to those of patients with KRAS mutant mCRC.
Conclusion: Gene expression appears to identify KRAS wt mCRC patients who receive little benefit from cetuximab. It will be important to test this model in an independent validation study.
Conflict of interest statement
The authors are employees of the respective companies listed.
Figures

 and += genes significantly associated with outcome, unadjusted P-value <0.05,
and += genes significantly associated with outcome, unadjusted P-value <0.05,  = genes associated with outcome controlling for FDR <0.05, *= genes not significantly associated with outcome. In total, 110 genes depicted. (B) Distribution of likelihood ratio P-values in KRAS wt patients by gene rank (objective response). Note:
= genes associated with outcome controlling for FDR <0.05, *= genes not significantly associated with outcome. In total, 110 genes depicted. (B) Distribution of likelihood ratio P-values in KRAS wt patients by gene rank (objective response). Note:  and += genes significantly associated with outcome, unadjusted P-value <0.05,
and += genes significantly associated with outcome, unadjusted P-value <0.05,  = genes associated with outcome controlling for FDR <0.05, *= genes not significantly associated with outcome. In total, 110 genes depicted. (C) Distribution of likelihood ratio P-values in KRAS wt patients by gene rank (PFS). Note:
= genes associated with outcome controlling for FDR <0.05, *= genes not significantly associated with outcome. In total, 110 genes depicted. (C) Distribution of likelihood ratio P-values in KRAS wt patients by gene rank (PFS). Note:  and += genes significantly associated with outcome, unadjusted P-value <0.05,
and += genes significantly associated with outcome, unadjusted P-value <0.05,  = genes associated with outcome controlling for FDR <0.05, *= genes not significantly associated with outcome. In total, 110 genes depicted.
= genes associated with outcome controlling for FDR <0.05, *= genes not significantly associated with outcome. In total, 110 genes depicted.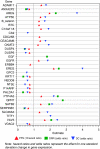
References
-
- Allegra CJ, Jessup JM, Somerfield MR, Hamilton SR, Hammond EH, Hayes DF, McAllister PK, Morton RF, Schilsky RL (2009) American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol 27: 2091–2096 - PubMed
-
- Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, Patterson SD, Chang DD (2008) Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol 26: 1626–1634 - PubMed
-
- Andreyev HJ, Norman AR, Cunningham D, Oates J, Dix BR, Iacopetta BJ, Young J, Walsh T, Ward R, Hawkins N, Beranek M, Jandik P, Benamouzig R, Jullian E, Laurent-Puig P, Olschwang S, Muller O, Hoffmann I, Rabes HM, Zietz C, Troungos C, Valavanis C, Yuen ST, Ho JW, Croke CT, O’Donoghue DP, Giaretti W, Rapallo A, Russo A, Bazan V, Tanaka M, Omura K, Azuma T, Ohkusa T, Fujimori T, Ono Y, Pauly M, Faber C, Glaesener R, de Goeij AF, Arends JW, Andersen SN, Lövig T, Breivik J, Gaudernack G, Clausen OP, De Angelis PD, Meling GI, Rognum TO, Smith R, Goh HS, Font A, Rosell R, Sun XF, Zhang H, Benhattar J, Losi L, Lee JQ, Wang ST, Clarke PA, Bell S, Quirke P, Bubb VJ, Piris J, Cruickshank NR, Morton D, Fox JC, Al-Mulla F, Lees N, Hall CN, Snary D, Wilkinson K, Dillon D, Costa J, Pricolo VE, Finkelstein SD, Thebo JS, Senagore AJ, Halter SA, Wadler S, Malik S, Krtolica K, Urosevic N (2001) Kirsten ras mutations in patients with colorectal cancer: the ‘RASCAL II’ study. Br J Cancer 85: 692–696 - PMC - PubMed
-
- Barber TD, Vogelstein B, Kinzler KW, Velculescu VE (2004) Somatic mutations of EGFR in colorectal cancers and glioblastomas. N Engl J Med 351: 2883. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

