Analyses of a set of 128 ancestry informative single-nucleotide polymorphisms in a global set of 119 population samples
- PMID: 21208434
- PMCID: PMC3025953
- DOI: 10.1186/2041-2223-2-1
Analyses of a set of 128 ancestry informative single-nucleotide polymorphisms in a global set of 119 population samples
Abstract
Background: Using DNA to determine an individual's ancestry from among human populations is generally interesting and useful for many purposes, including admixture mapping, controlling for population structure in disease or trait association studies and forensic ancestry inference. However, to estimate ancestry, including possible admixture within an individual, as well as heterogeneity within a group of individuals, allele frequencies are necessary for what are believed to be the contributing populations. For this purpose, panels of ancestry informative markers (AIMs) have been developed.
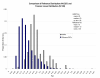
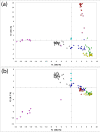
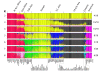
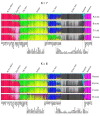
Results: We are presenting our work on one such panel, composed of 128 ancestry informative single-nucleotide polymorphisms (AISNPs) already proposed in the literature. Compared to previous studies of these AISNPs, we have studied three times the number of individuals (4,871) in three times as many population samples (119). We have validated this panel for many ancestry assignment and admixture studies, especially those that were the rationale for the original selection of the 128 SNPs: African Americans and Mexican Americans. At the same time, the limitations of the panel for distinguishing ancestry and quantifying admixture among Eurasian populations are noted.
Conclusion: We demonstrate the simultaneous importance of the specific set of population samples and their relative sample sizes in the use of the structure program to determine which groups cluster together and consequently influence the ability of a marker panel to infer ancestry. We demonstrate the strengths and weaknesses of this particular panel of AISNPs in a global context.
Figures

References
-
- Bryc K, Auton A, Nelson MR, Oksenberg JR, Hauser SL, Williams S, Froment A, Bodo JM, Wambebe C, Tishkoff SA, Bustamante CD. Genome-wide patterns of population structure and admixture in West Africans and African Americans. Proc Natl Acad Sci USA. 2010;107:786–791. doi: 10.1073/pnas.0909559107. - DOI - PMC - PubMed
-
- Price AL, Patterson N, Yu F, Cox DR, Waliszewska A, McDonald GJ, Tandon A, Schirmer C, Neubauer J, Bedoya G, Duque C, Villegas A, Bortolini MC, Salzano FM, Gallo C, Mazzotti G, Tello-Ruiz M, Riba L, Aguilar-Salinas CA, Canizales-Quinteros S, Menjivar M, Klitz W, Henderson B, Haiman CA, Winkler C, Tusie-Luna T, Ruiz-Linares A, Reich D. A genomewide admixture map for Latino populations. Am J Hum Genet. 2007;80:1024–1036. doi: 10.1086/518313. - DOI - PMC - PubMed
-
- Santos NP, Ribeiro-Rodrigues EM, Ribeiro-Dos-Santos AK, Pereira R, Gusmão L, Amorim A, Guerreiro JF, Zago MA, Matte C, Hutz MH, Santos SE. Assessing individual interethnic admixture and population substructure using a 48-insertion-deletion (INSEL) ancestry-informative marker (AIM) panel. Hum Mutat. 2010;31:184–190. doi: 10.1002/humu.21159. - DOI - PubMed
-
- Lins TC, Vieira RG, Abreu BS, Grattapaglia D, Pereira RW. Genetic composition of Brazilian population samples based on a set of twenty-eight ancestry informative SNPs. Am J Hum Biol. 2010;22:187–192. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

