The genome sequence of E. coli W (ATCC 9637): comparative genome analysis and an improved genome-scale reconstruction of E. coli
- PMID: 21208457
- PMCID: PMC3032704
- DOI: 10.1186/1471-2164-12-9
The genome sequence of E. coli W (ATCC 9637): comparative genome analysis and an improved genome-scale reconstruction of E. coli
Abstract
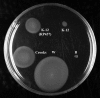
Background: Escherichia coli is a model prokaryote, an important pathogen, and a key organism for industrial biotechnology. E. coli W (ATCC 9637), one of four strains designated as safe for laboratory purposes, has not been sequenced. E. coli W is a fast-growing strain and is the only safe strain that can utilize sucrose as a carbon source. Lifecycle analysis has demonstrated that sucrose from sugarcane is a preferred carbon source for industrial bioprocesses.
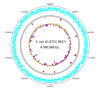
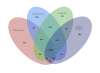
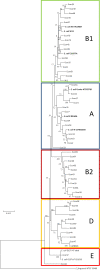
Results: We have sequenced and annotated the genome of E. coli W. The chromosome is 4,900,968 bp and encodes 4,764 ORFs. Two plasmids, pRK1 (102,536 bp) and pRK2 (5,360 bp), are also present. W has unique features relative to other sequenced laboratory strains (K-12, B and Crooks): it has a larger genome and belongs to phylogroup B1 rather than A. W also grows on a much broader range of carbon sources than does K-12. A genome-scale reconstruction was developed and validated in order to interrogate metabolic properties.
Conclusions: The genome of W is more similar to commensal and pathogenic B1 strains than phylogroup A strains, and therefore has greater utility for comparative analyses with these strains. W should therefore be the strain of choice, or 'type strain' for group B1 comparative analyses. The genome annotation and tools created here are expected to allow further utilization and development of E. coli W as an industrial organism for sucrose-based bioprocesses. Refinements in our E. coli metabolic reconstruction allow it to more accurately define E. coli metabolism relative to previous models.
Figures
References
-
- Gunsalus IC, Hand DB. The use of bacteria in the chemical determination of total vitamin C. J Biol Chem. 1941;141(3):853–858.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

