The molecular evolution of the p120-catenin subfamily and its functional associations
- PMID: 21209830
- PMCID: PMC3013132
- DOI: 10.1371/journal.pone.0015747
The molecular evolution of the p120-catenin subfamily and its functional associations
Abstract
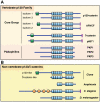
Background: p120-catenin (p120) is the prototypical member of a subclass of armadillo-related proteins that includes δ-catenin/NPRAP, ARVCF, p0071, and the more distantly related plakophilins 1-3. In vertebrates, p120 is essential in regulating surface expression and stability of all classical cadherins, and directly interacts with Kaiso, a BTB/ZF family transcription factor.
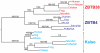
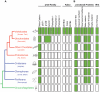
Methodology/principal findings: To clarify functional relationships between these proteins and how they relate to the classical cadherins, we have examined the proteomes of 14 diverse vertebrate and metazoan species. The data reveal a single ancient δ-catenin-like p120 family member present in the earliest metazoans and conserved throughout metazoan evolution. This single p120 family protein is present in all protostomes, and in certain early-branching chordate lineages. Phylogenetic analyses suggest that gene duplication and functional diversification into "p120-like" and "δ-catenin-like" proteins occurred in the urochordate-vertebrate ancestor. Additional gene duplications during early vertebrate evolution gave rise to the seven vertebrate p120 family members. Kaiso family members (i.e., Kaiso, ZBTB38 and ZBTB4) are found only in vertebrates, their origin following that of the p120-like gene lineage and coinciding with the evolution of vertebrate-specific mechanisms of epigenetic gene regulation by CpG island methylation.
Conclusions/significance: The p120 protein family evolved from a common δ-catenin-like ancestor present in all metazoans. Through several rounds of gene duplication and diversification, however, p120 evolved in vertebrates into an essential, ubiquitously expressed protein, whereas loss of the more selectively expressed δ-catenin, p0071 and ARVCF are tolerated in most species. Together with phylogenetic studies of the vertebrate cadherins, our data suggest that the p120-like and δ-catenin-like genes co-evolved separately with non-neural (E- and P-cadherin) and neural (N- and R-cadherin) cadherin lineages, respectively. The expansion of p120 relative to δ-catenin during vertebrate evolution may reflect the pivotal and largely disproportionate role of the non-neural cadherins with respect to evolution of the wide range of somatic morphology present in vertebrates today.
Conflict of interest statement
Figures

References
-
- Schneider SQ, Finnerty JR, Martindale MQ. Protein evolution: structure-function relationships of the oncogene beta-catenin in the evolution of multicellular animals. J Exp Zool B Mol Dev Evol. 2003;295:25–44. - PubMed
-
- Gallin WJ. Evolution of the “classical” cadherin family of cell adhesion molecules in vertebrates. Mol Biol Evol. 1998;15:1099–1107. - PubMed
-
- Hulpiau P, van Roy F. Molecular evolution of the cadherin superfamily. Int J Biochem Cell Biol. 2009;41:349–369. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

