Dynamics of glycoprotein charge in the evolutionary history of human influenza
- PMID: 21209885
- PMCID: PMC3012697
- DOI: 10.1371/journal.pone.0015674
Dynamics of glycoprotein charge in the evolutionary history of human influenza
Abstract
Background: Influenza viruses show a significant capacity to evade host immunity; this is manifest both as large occasional jumps in the antigenic phenotype of viral surface molecules and in gradual antigenic changes leading to annual influenza epidemics in humans. Recent mouse studies show that avidity for host cells can play an important role in polyclonal antibody escape, and further that electrostatic charge of the hemagglutinin glycoprotein can contribute to such avidity.
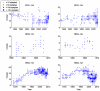
Methodology/principal findings: We test the role of glycoprotein charge on sequence data from the three major subtypes of influenza A in humans, using a simple method of calculating net glycoprotein charge. Of all subtypes, H3N2 in humans shows a striking pattern of increasing positive charge since its introduction in 1968. Notably, this trend applies to both hemagglutinin and neuraminidase glycoproteins. In the late 1980s hemagglutinin charge reached a plateau, while neuraminidase charge started to decline. We identify key groups of amino acid sites involved in this charge trend.
Conclusions/significance: To our knowledge these are the first indications that, for human H3N2, net glycoprotein charge covaries strongly with antigenic drift on a global scale. Further work is needed to elucidate how such charge interacts with other immune escape mechanisms, such as glycosylation, and we discuss important questions arising for future study.
Conflict of interest statement
Figures
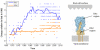

References
-
- Grenfell BT, Pybus OB, Gog JR, Wood JLN, Daley JM, et al. Unifying the epidemiological and evolutionary dynamics of pathogens. Science. 2004;Jan 16; 303(5656):327–32. - PubMed
-
- Carrat F, Flahault A. Influenza vaccine: the challenge of antigenic drift. Vaccine Sep 28;25(39-40):6852-62. Epub. 2007;2007 Aug 3 - PubMed
-
- Webster RG, Laver WG, Air GM, Schild GC. Molecular mechanisms of variation in influenza viruses. Nature. 1982;Mar 11; 296(5853):115–21. - PubMed
-
- Smith DJ, Lapedes AS, de Jong JC, Bestebroer TM, Rimmelzwaan GF, et al. Mapping the antigenic and genetic evolution of influenza virus. Science Jul 16;305(5682):371-6. Epub. 2004;2004 Jun 24 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

