A complete analysis of HA and NA genes of influenza A viruses
- PMID: 21209922
- PMCID: PMC3012125
- DOI: 10.1371/journal.pone.0014454
A complete analysis of HA and NA genes of influenza A viruses
Abstract
Background: More and more nucleotide sequences of type A influenza virus are available in public databases. Although these sequences have been the focus of many molecular epidemiological and phylogenetic analyses, most studies only deal with a few representative sequences. In this paper, we present a complete analysis of all Haemagglutinin (HA) and Neuraminidase (NA) gene sequences available to allow large scale analyses of the evolution and epidemiology of type A influenza.
Methodology/principal findings: This paper describes an analysis and complete classification of all HA and NA gene sequences available in public databases using multivariate and phylogenetic methods.
Conclusions/significance: We analyzed 18,975 HA sequences and divided them into 280 subgroups according to multivariate and phylogenetic analyses. Similarly, we divided 11,362 NA sequences into 202 subgroups. Compared to previous analyses, this work is more detailed and comprehensive, especially for the bigger datasets. Therefore, it can be used to show the full and complex phylogenetic diversity and provides a framework for studying the molecular evolution and epidemiology of type A influenza virus. For more than 85% of type A influenza HA and NA sequences into GenBank, they are categorized in one unambiguous and unique group. Therefore, our results are a kind of genetic and phylogenetic annotation for influenza HA and NA sequences. In addition, sequences of swine influenza viruses come from 56 HA and 45 NA subgroups. Most of these subgroups also include viruses from other hosts indicating cross species transmission of the viruses between pigs and other hosts. Furthermore, the phylogenetic diversity of swine influenza viruses from Eurasia is greater than that of North American strains and both of them are becoming more diverse. Apart from viruses from human, pigs, birds and horses, viruses from other species show very low phylogenetic diversity. This might indicate that viruses have not become established in these species. Based on current evidence, there is no simple pattern of inter-hemisphere transmission of avian influenza viruses and it appears to happen sporadically. However, for H6 subtype avian influenza viruses, such transmissions might have happened very frequently and multiple and bidirectional transmission events might exist.
Conflict of interest statement
Figures
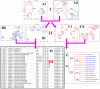

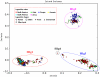

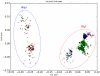


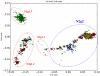
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

