The enigmatic origin of bovine mtDNA haplogroup R: sporadic interbreeding or an independent event of Bos primigenius domestication in Italy?
- PMID: 21209945
- PMCID: PMC3011016
- DOI: 10.1371/journal.pone.0015760
The enigmatic origin of bovine mtDNA haplogroup R: sporadic interbreeding or an independent event of Bos primigenius domestication in Italy?
Abstract
Background: When domestic taurine cattle diffused from the Fertile Crescent, local wild aurochsen (Bos primigenius) were still numerous. Moreover, aurochsen and introduced cattle often coexisted for millennia, thus providing potential conditions not only for spontaneous interbreeding, but also for pastoralists to create secondary domestication centers involving local aurochs populations. Recent mitochondrial genomes analyses revealed that not all modern taurine mtDNAs belong to the shallow macro-haplogroup T of Near Eastern origin, as demonstrated by the detection of three branches (P, Q and R) radiating prior to the T node in the bovine phylogeny. These uncommon haplogroups represent excellent tools to evaluate if sporadic interbreeding or even additional events of cattle domestication occurred.
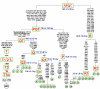
Methodology: The survey of the mitochondrial DNA (mtDNA) control-region variation of 1,747 bovine samples (1,128 new and 619 from previous studies) belonging to 37 European breeds allowed the identification of 16 novel non-T mtDNAs, which after complete genome sequencing were confirmed as members of haplogroups Q and R. These mtDNAs were then integrated in a phylogenetic tree encompassing all available P, Q and R complete mtDNA sequences.
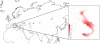
Conclusions: Phylogenetic analyses of 28 mitochondrial genomes belonging to haplogroups P (N = 2), Q (N = 16) and R (N = 10) together with an extensive survey of all previously published mtDNA datasets revealed major similarities between haplogroups Q and T. Therefore, Q most likely represents an additional minor lineage domesticated in the Near East together with the founders of the T subhaplogroups. Whereas, haplogroup R is found, at least for the moment, only in Italy and nowhere else, either in modern or ancient samples, thus supporting an origin from European aurochsen. Haplogroup R could have been acquired through sporadic interbreeding of wild and domestic animals, but our data do not rule out the possibility of a local and secondary event of B. primigenius domestication in Italy.
Conflict of interest statement
Figures

References
-
- Mason H. Evolution of domestic animals. London: Longman. 1984;xii:452.
-
- Clutton-Brock J. The walking larder: patterns of domestication, pastoralism and predation. London: Unwin Hyamn. 1989;xxii:368.
-
- Meadow RH. Possehl G, editor. in Harappan civilisation, 2nd edn. pp. 295–320. (Oxford & IBH, New Delhi, India, 1993)
-
- Troy CS, MacHugh DE, Bailey JF, Magee DA, Loftus RT, et al. Genetic evidence for Near-Eastern origins of European cattle. Nature. 2001;410:1088–1099. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

