Formation and toxicity of soluble polyglutamine oligomers in living cells
- PMID: 21209946
- PMCID: PMC3011017
- DOI: 10.1371/journal.pone.0015245
Formation and toxicity of soluble polyglutamine oligomers in living cells
Abstract
Background: Aggregation and cytotoxicity of mutant proteins containing an expanded number of polyglutamine (polyQ) repeats is a hallmark of several diseases, including Huntington's disease (HD). Within cells, mutant Huntingtin (mHtt) and other polyglutamine expansion mutant proteins exist as monomers, soluble oligomers, and insoluble inclusion bodies (IBs). Determining which of these forms constitute a toxic species has proven difficult. Recent studies support a role for IBs as a cellular coping mechanism to sequester levels of potentially toxic soluble monomeric and oligomeric species of mHtt.
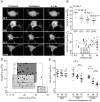
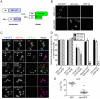
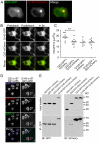
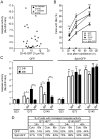
Methodology/principal findings: When fused to a fluorescent reporter (GFP) and expressed in cells, the soluble monomeric and oligomeric polyglutamine species are visually indistinguishable. Here, we describe two complementary biophysical fluorescence microscopy techniques to directly detect soluble polyglutamine oligomers (using Htt exon 1 or Htt(ex1)) and monitor their fates in live cells. Photobleaching analyses revealed a significant reduction in the mobilities of mHtt(ex1) variants consistent with their incorporation into soluble microcomplexes. Similarly, when fused to split-GFP constructs, both wildtype and mHtt(ex1) formed oligomers, as evidenced by the formation of a fluorescent reporter. Only the mHtt(ex1) split-GFP oligomers assembled into IBs. Both FRAP and split-GFP approaches confirmed the ability of mHtt(ex1) to bind and incorporate wildtype Htt into soluble oligomers. We exploited the irreversible binding of split-GFP fragments to forcibly increase levels of soluble oligomeric mHtt(ex1). A corresponding increase in the rate of IBs formation and the number formed was observed. Importantly, higher levels of soluble mHtt(ex1) oligomers significantly correlated with increased mutant cytotoxicity, independent of the presence of IBs.
Conclusions/significance: Our study describes powerful and sensitive tools for investigating soluble oligomeric forms of expanded polyglutamine proteins, and their impact on cell viability. Moreover, these methods should be applicable for the detection of soluble oligomers of a wide variety of aggregation prone proteins.
Conflict of interest statement
Figures



References
-
- Lippincott-Schwartz J, Snapp E, Kenworthy A. Studying protein dynamics in living cells. Nat Rev Mol Cell Biol. 2001;2:444–456. - PubMed
-
- Wouters FS, Bastiaens PI. Imaging protein-protein interactions by fluorescence resonance energy transfer (FRET) microscopy. Curr Protoc Protein Sci Chapter. 2001;19:Unit19 15. - PubMed
-
- Villalobos V, Naik S, Piwnica-Worms D. Detection of protein-protein interactions in live cells and animals with split firefly luciferase protein fragment complementation. Methods Mol Biol. 2008;439:339–352. - PubMed
-
- Gusella JF, MacDonald ME. Huntington's disease: seeing the pathogenic process through a genetic lens. Trends Biochem Sci. 2006;31:533–540. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

