The SOX2 response program in glioblastoma multiforme: an integrated ChIP-seq, expression microarray, and microRNA analysis
- PMID: 21211035
- PMCID: PMC3022822
- DOI: 10.1186/1471-2164-12-11
The SOX2 response program in glioblastoma multiforme: an integrated ChIP-seq, expression microarray, and microRNA analysis
Abstract
Background: SOX2 is a key gene implicated in maintaining the stemness of embryonic and adult stem cells. SOX2 appears to re-activate in several human cancers including glioblastoma multiforme (GBM), however, the detailed response program of SOX2 in GBM has not yet been defined.
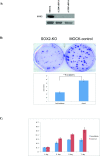
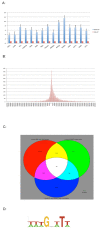
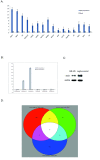
Results: We show that knockdown of the SOX2 gene in LN229 GBM cells reduces cell proliferation and colony formation. We then comprehensively characterize the SOX2 response program by an integrated analysis using several advanced genomic technologies including ChIP-seq, microarray profiling, and microRNA sequencing. Using ChIP-seq technology, we identified 4883 SOX2 binding regions in the GBM cancer genome. SOX2 binding regions contain the consensus sequence wwTGnwTw that occurred 3931 instances in 2312 SOX2 binding regions. Microarray analysis identified 489 genes whose expression altered in response to SOX2 knockdown. Interesting findings include that SOX2 regulates the expression of SOX family proteins SOX1 and SOX18, and that SOX2 down regulates BEX1 (brain expressed X-linked 1) and BEX2 (brain expressed X-linked 2), two genes with tumor suppressor activity in GBM. Using next generation sequencing, we identified 105 precursor microRNAs (corresponding to 95 mature miRNAs) regulated by SOX2, including down regulation of miR-143, -145, -253-5p and miR-452. We also show that miR-145 and SOX2 form a double negative feedback loop in GBM cells, potentially creating a bistable system in GBM cells.
Conclusions: We present an integrated dataset of ChIP-seq, expression microarrays and microRNA sequencing representing the SOX2 response program in LN229 GBM cells. The insights gained from our integrated analysis further our understanding of the potential actions of SOX2 in carcinogenesis and serves as a useful resource for the research community.
Figures

References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

