Chromodomain-mediated oligomerization of HP1 suggests a nucleosome-bridging mechanism for heterochromatin assembly
- PMID: 21211724
- PMCID: PMC3752404
- DOI: 10.1016/j.molcel.2010.12.016
Chromodomain-mediated oligomerization of HP1 suggests a nucleosome-bridging mechanism for heterochromatin assembly
Abstract
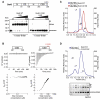
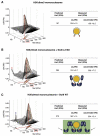
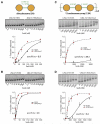
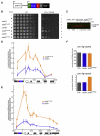
HP1 proteins are central to the assembly and spread of heterochromatin containing histone H3K9 methylation. The chromodomain (CD) of HP1 proteins specifically recognizes the methyl mark on H3 peptides, but the same extent of specificity is not observed within chromatin. The chromoshadow domain of HP1 proteins promotes homodimerization, but this alone cannot explain heterochromatin spread. Using the S. pombe HP1 protein, Swi6, we show that recognition of H3K9-methylated chromatin in vitro relies on an interface between two CDs. This interaction causes Swi6 to tetramerize on a nucleosome, generating two vacant CD sticky ends. On nucleosomal arrays, methyl mark recognition is highly sensitive to internucleosomal distance, suggesting that the CD sticky ends bridge nearby methylated nucleosomes. Strengthening the CD-CD interaction enhances silencing and heterochromatin spread in vivo. Our findings suggest that recognition of methylated nucleosomes and HP1 spread on chromatin are structurally coupled and imply that methylation and nucleosome arrangement synergistically regulate HP1 function.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures



References
-
- Cowieson NP, Partridge JF, Allshire RC, McLaughlin PJ. Dimerisation of a chromo shadow domain and distinctions from the chromodomain as revealed by structural analysis. Curr Biol. 2000;10:517–525. - PubMed
-
- Danzer JR, Wallrath LL. Mechanisms of HP1-mediated gene silencing in Drosophila. Development. 2004;131:3571–3580. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

