Reprogramming factor expression initiates widespread targeted chromatin remodeling
- PMID: 21211784
- PMCID: PMC3220622
- DOI: 10.1016/j.stem.2010.12.001
Reprogramming factor expression initiates widespread targeted chromatin remodeling
Abstract
Despite rapid progress in characterizing transcription factor-driven reprogramming of somatic cells to an induced pluripotent stem cell (iPSC) state, many mechanistic questions still remain. To gain insight into the earliest events in the reprogramming process, we systematically analyzed the transcriptional and epigenetic changes that occur during early factor induction after discrete numbers of divisions. We observed rapid, genome-wide changes in the euchromatic histone modification, H3K4me2, at more than a thousand loci including large subsets of pluripotency-related or developmentally regulated gene promoters and enhancers. In contrast, patterns of the repressive H3K27me3 modification remained largely unchanged except for focused depletion specifically at positions where H3K4 methylation is gained. These chromatin regulatory events precede transcriptional changes within the corresponding loci. Our data provide evidence for an early, organized, and population-wide epigenetic response to ectopic reprogramming factors that clarify the temporal order through which somatic identity is reset during reprogramming.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures

Schematic for enrichment of distinct proliferative cohorts using the live dye CFSE and serum pulsing under constant factor induction and time. After 96 hours of continued culture in doxycycline-supplemented medium, samples were sorted and scored using Flow Cytometry. Median fluorophore intensity was assessed as a relative metric for proliferative number and is shown on the right. Relative intensity is displayed in arbitrary units (A.U.).
mRNA expression dynamics conditional on MEF/ES chromatin state progressing across cell division number (shown color coded in the inset) for up- and down-regulated genes. ES cell H3K4me3 only loci and their respective states in MEFs are shown on the left and ES cell bivalent (H3K4me3/H3K27me3) loci are shown on the right.
Enrichment for Oct4, Sox2, Klf4 and c-Myc (OSKM) binding in promoter elements of dynamically regulated genes shows an asymmetric bias towards gene activation within targets of the myc oncogene. Transcription factor binding taken from genome-scale profiling of embryonic stem cells (Kim et al., 2008; Marson et al., 2008).
Density plot of genes with dynamic H3K4me2 in reprogramming populations compared to control MEFs. Promoters exhibiting a dynamic shift in H3K4me2 (n~1,500) fall into 3 distinct classes: de novo (beige), enhanced (red) and loss (green). Representative genes from all three classes are highlighted on the right.
Expression data between starting state (control) and the >3 divisions induced population with dynamic H3K4me2 genes highlighted in red. Pie chart shows the representation of genes that exhibit only H3K4me2 changes (pink) or both H3K4me2 and gene expression changes (red; n~10%).
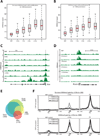
De novo H3K4me2 acquisition is continuous across cohorts and already visible before a single division (n~300). Red line indicates median. Whiskers represent 2.5 and 97.5 percentile.
Enhanced H3K4me2 at a subset of ~1000 promoters over proliferative cohorts exhibit similar trends and approach expected ES cell levels in dividing populations of reprogramming cells. Red line indicates median. Whiskers represent 2.5 and 97.5 percentile.
ChIP-Seq tracks showing de novo H3K4me2 at the endogenous promoter of Aire as part of an orchestrated enrichment that is preferential for Oct4 and Sox2 regulated promoters. Green bars on the bottom indicate the CpG island to which H3K4 methylation is restricted. Gray bar highlights the nucleosome depleted region that is flanked by H3K4me2 within ES cells.
H3K4me2 ChiP-seq map of the Postn locus, which is expressed in MEFs and silenced by >3 divisions, shows a loss of H3K4me2 levels at its promoter region to ES cell like levels. The Postn locus represents 115 promoters for which H3K4me2 is lost during reprogramming factor induction.
ES cell transcription factor occupancy of genes demonstrating H3K4me2 enrichment show a predominance of Oct4 and Sox2 binding.
Composite plots of H3K4 mono-, di-, and trimethylation distribution at de novo and enhanced promoter classes in control MEFs, after 3 divisions, and within ES cells. In the de novo set (n~300 genes) H3K4me2 gain is exclusive, whereas concurrent enrichment for H3K4me2 and H3K4me3 is present at enhanced promoters (n~1400 genes) in which H3K4 methylation increases at least 2.5 fold..
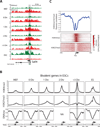
The Sall4 locus exhibits a de novo gain of H3K4 methylation at two CpG Islands (green bars). Gain of H3K4me2 corresponds to a targeted depletion of H3K27 methylation within cycling cells that is limited to the site of H3K4 methylation. Highlighted region displays the CpG island and the site of ES cell specific nucleosome depletion.
B. General trends of epigenetic reprogramming events at ES bivalent promoters (n=688) within induced populations:
Upper Panel: Composite plots of H3K4me2 gain within ES cell bivalent promoters compared against somatic and ES cell controls.
Middle Panel: Composite plot of H3K27me3 levels stay constant except in the most proliferative cohort (>3 divisions) where levels are inversely proportional to the gain in H3K4me2 and are subsequently depleted.
Lower Panel: CpG methylation values at regions of enhanced H3K4me2 gain are predominantly hypomethylated across states as expected given the high CpG density of this promoter set (82% CpG islands).
CpG density across the promoters analyzed is highlighted and demonstrates the boundary of the dynamic changes in chromatin state. Scale ranges between 40% (white) and 80% (black) GC content.
Pearson correlation between H3K4me2 and H3K27me3 levels in 200 base pair sliding windows. Negative correlation between the two marks reaches significance within 500 bp from the TSS. Histone mark enrichments for the promoter set are included as heat maps and emphasize this inverse relationship.
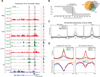
The CpG Island promoter (P, red highlight) of the ES cell expressed st14 gene displays minimal H3K4 methylation in the somatic state and increases in H3K4me2 with proliferation, concurrent with punctate loss of H3K27me3 at the CpG Island (see also Figure 3A). The de novo K4me2 gain is accompanied by gain of an intronic enhancer signature (E, red highlight). Expression levels for st14 are not detected until complete remodeling at later stages. Intergenic enhancers (E, red highlight, right) are also gained and are progressively enriched for H3K4me1 and me2.
Number of MEF exclusive or ES exclusive putative enhancers that are gained or lost across division. The “ES specific” enhancer set does not include the 3,708 enhancers that are shared between MEF, ES cells, and all reprogramming populations. Inset: Venn diagram of represented enhancers within reprogramming cells against the starting somatic state and ES cells.
Architecture and relationship of H3K4 methylation marks gained at newly acquired enhancer signatures called after >3 divisions as in (B). Enhancers gain significant H3K4me1 in early proliferative cohorts followed by subsequent H3K4me2 enrichment.
Composite plot of ES cell H3K4me2 enhancer peaks gained in reprogramming populations demonstrate an equivalent CpG hypomethylation in somatic and ES cells. Alternatively, ES cell specific enhancers that are not acquired after 96 hours of factor induction demonstrate differential and higher mean CpG methylation. Dashed lines highlight somatic CpG methylation in the acquired versus ES cell exclusive sets.
References
-
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, 3rd, Gingeras TR, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. - PubMed
-
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. - PubMed
-
- Boland MJ, Hazen JL, Nazor KL, Rodriguez AR, Gifford W, Martin G, Kupriyanov S, Baldwin KK. Adult mice generated from induced pluripotent stem cells. Nature. 2009;461:91–94. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

