Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons
- PMID: 21212162
- PMCID: PMC3044863
- DOI: 10.1101/gr.112730.110
Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons
Abstract
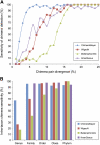
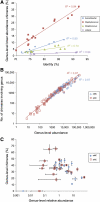
Bacterial diversity among environmental samples is commonly assessed with PCR-amplified 16S rRNA gene (16S) sequences. Perceived diversity, however, can be influenced by sample preparation, primer selection, and formation of chimeric 16S amplification products. Chimeras are hybrid products between multiple parent sequences that can be falsely interpreted as novel organisms, thus inflating apparent diversity. We developed a new chimera detection tool called Chimera Slayer (CS). CS detects chimeras with greater sensitivity than previous methods, performs well on short sequences such as those produced by the 454 Life Sciences (Roche) Genome Sequencer, and can scale to large data sets. By benchmarking CS performance against sequences derived from a controlled DNA mixture of known organisms and a simulated chimera set, we provide insights into the factors that affect chimera formation such as sequence abundance, the extent of similarity between 16S genes, and PCR conditions. Chimeras were found to reproducibly form among independent amplifications and contributed to false perceptions of sample diversity and the false identification of novel taxa, with less-abundant species exhibiting chimera rates exceeding 70%. Shotgun metagenomic sequences of our mock community appear to be devoid of 16S chimeras, supporting a role for shotgun metagenomics in validating novel organisms discovered in targeted sequence surveys.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
- U54-HG004969/HG/NHGRI NIH HHS/United States
- U54-HG003079/HG/NHGRI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- U54 AI084844/AI/NIAID NIH HHS/United States
- U54 HG003273/HG/NHGRI NIH HHS/United States
- U01 HG004866/HG/NHGRI NIH HHS/United States
- N01-AI30071/AI/NIAID NIH HHS/United States
- U54-AI084844/AI/NIAID NIH HHS/United States
- U01-HG004866/HG/NHGRI NIH HHS/United States
- U54 HG003079/HG/NHGRI NIH HHS/United States
- U54 HG004969/HG/NHGRI NIH HHS/United States
- U54-HG004973/HG/NHGRI NIH HHS/United States
- N01 AI030071/AI/NIAID NIH HHS/United States
- U54-HG003273/HG/NHGRI NIH HHS/United States
- U54 HG004973/HG/NHGRI NIH HHS/United States
- U54-HG004968/HG/NHGRI NIH HHS/United States
- U54 HG004968/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
