Kaposi sarcoma-associated herpesvirus degrades cellular Toll-interleukin-1 receptor domain-containing adaptor-inducing beta-interferon (TRIF)
- PMID: 21212282
- PMCID: PMC3048673
- DOI: 10.1074/jbc.M110.191452
Kaposi sarcoma-associated herpesvirus degrades cellular Toll-interleukin-1 receptor domain-containing adaptor-inducing beta-interferon (TRIF)
Abstract
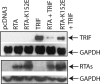
Kaposi sarcoma-associated herpesvirus (KSHV) is a human γ-herpesvirus associated with several human malignancies. The replication and transcription activator (RTA) is necessary and sufficient for the switch from KSHV latency to lytic replication. Toll-interleukin-1 receptor (TIR) domain-containing adaptor-inducing β-interferon (TRIF, also called TIR-domain-containing adaptor molecule-1 (TICAM-1)) is a signaling adaptor molecule that is critically involved in the Toll-like receptor 3 (TLR-3) and TLR-4 signaling pathways for type I interferon (IFN) production, a key component of innate immunity against microbial infection. In this report, we find a new mechanism by which RTA blocks innate immunity by targeting cellular TRIF. RTA specifically degrades TRIF by shortening the half-life of TRIF protein. This RTA-mediated degradation is at least partially mediated through the ubiquitin-proteasome pathway because proteasome inhibitors as well as knockdown of cellular ubiquitin expression alleviate the degradation. RTA may not directly interact with TRIF and may activate TRIF degradation indirectly through an unknown mediator(s). RTA targets multiple regions of TRIF and may use its ubiquitin ligase domain for the degradation. In addition, physiological levels of TRIF protein are down-regulated during KSHV lytic replication when RTA is expressed. Finally, RTA down-regulates double-stranded RNA-initiated activation of TLR-3 pathway, in the absence of degradation of IFN regulatory factor 7 (IRF-7). Taken together, these data suggest that KSHV employs a novel mechanism to block the innate immunity by degrading TRIF protein. This work may contribute to our understandings on how KSHV evades host immunity for its survival in vivo.
Figures
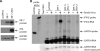


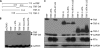
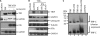


References
-
- Moore P., Chang Y. (2001) in Virology (Knipe D. M. ed) pp. 2803–2833, Lippincott Williams and Wilkins, Philadelphia, PA
-
- Chang Y., Cesarman E., Pessin M. S., Lee F., Culpepper J., Knowles D. M., Moore P. S. (1994) Science 266, 1865–1869 - PubMed
-
- Chang Y., Moore P. S. (1996) Infect. Agents Dis. 5, 215–222 - PubMed
-
- West J. T., Wood C. (2003) Oncogene 22, 5150–5163 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

