Suppression of leukemia development caused by PTEN loss
- PMID: 21212363
- PMCID: PMC3029702
- DOI: 10.1073/pnas.1006937108
Suppression of leukemia development caused by PTEN loss
Abstract
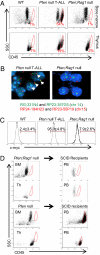
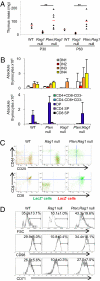
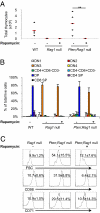
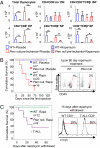
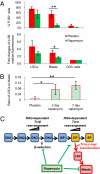
Multiple genetic or molecular alterations are known to be associated with cancer stem cell formation and cancer development. Targeting such alterations, therefore, may lead to cancer prevention. By crossing our previously established phosphatase and tensin homolog (Pten)-null acute T-lymphoblastic leukemia (T-ALL) model onto the recombination-activating gene 1(-/-) background, we show that the lack of variable, diversity and joining [V(D)J] recombination completely abolishes the Tcrα/δ-c-myc translocation and T-ALL development, regardless of β-catenin activation. We identify mammalian target of rapamycin (mTOR) as a regulator of β-selection. Rapamycin, an mTOR-specific inhibitor, alters nutrient sensing and blocks T-cell differentiation from CD4(-)CD8(-) to CD4(+)CD8(+), the stage where the Tcrα/δ-c-myc translocation occurs. Long-term rapamycin treatment of preleukemic Pten-null mice prevents Tcrα/δ-c-myc translocation and leukemia stem cell (LSC) formation, and it halts T-ALL development. However, rapamycin alone fails to inhibit mTOR signaling in the c-Kit(mid)CD3(+)Lin(-) population enriched for LSCs and eliminate these cells. Our results support the idea that preventing LSC formation and selectively targeting LSCs are promising approaches for antileukemia therapies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

