A structure-based benchmark for protein-protein binding affinity
- PMID: 21213247
- PMCID: PMC3064828
- DOI: 10.1002/pro.580
A structure-based benchmark for protein-protein binding affinity
Abstract
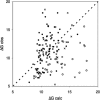
We have assembled a nonredundant set of 144 protein-protein complexes that have high-resolution structures available for both the complexes and their unbound components, and for which dissociation constants have been measured by biophysical methods. The set is diverse in terms of the biological functions it represents, with complexes that involve G-proteins and receptor extracellular domains, as well as antigen/antibody, enzyme/inhibitor, and enzyme/substrate complexes. It is also diverse in terms of the partners' affinity for each other, with K(d) ranging between 10(-5) and 10(-14) M. Nine pairs of entries represent closely related complexes that have a similar structure, but a very different affinity, each pair comprising a cognate and a noncognate assembly. The unbound structures of the component proteins being available, conformation changes can be assessed. They are significant in most of the complexes, and large movements or disorder-to-order transitions are frequently observed. The set may be used to benchmark biophysical models aiming to relate affinity to structure in protein-protein interactions, taking into account the reactants and the conformation changes that accompany the association reaction, instead of just the final product.
Copyright © 2011 The Protein Society.
Figures
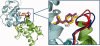
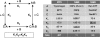
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

