Building prognostic models for breast cancer patients using clinical variables and hundreds of gene expression signatures
- PMID: 21214954
- PMCID: PMC3025826
- DOI: 10.1186/1755-8794-4-3
Building prognostic models for breast cancer patients using clinical variables and hundreds of gene expression signatures
Abstract
Background: Multiple breast cancer gene expression profiles have been developed that appear to provide similar abilities to predict outcome and may outperform clinical-pathologic criteria; however, the extent to which seemingly disparate profiles provide additive prognostic information is not known, nor do we know whether prognostic profiles perform equally across clinically defined breast cancer subtypes. We evaluated whether combining the prognostic powers of standard breast cancer clinical variables with a large set of gene expression signatures could improve on our ability to predict patient outcomes.
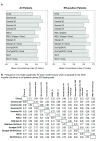
Methods: Using clinical-pathological variables and a collection of 323 gene expression "modules", including 115 previously published signatures, we build multivariate Cox proportional hazards models using a dataset of 550 node-negative systemically untreated breast cancer patients. Models predictive of pathological complete response (pCR) to neoadjuvant chemotherapy were also built using this approach.
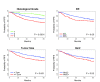
Results: We identified statistically significant prognostic models for relapse-free survival (RFS) at 7 years for the entire population, and for the subgroups of patients with ER-positive, or Luminal tumors. Furthermore, we found that combined models that included both clinical and genomic parameters improved prognostication compared with models with either clinical or genomic variables alone. Finally, we were able to build statistically significant combined models for pathological complete response (pCR) predictions for the entire population.
Conclusions: Integration of gene expression signatures and clinical-pathological factors is an improved method over either variable type alone. Highly prognostic models could be created when using all patients, and for the subset of patients with lymph node-negative and ER-positive breast cancers. Other variables beyond gene expression and clinical-pathological variables, like gene mutation status or DNA copy number changes, will be needed to build robust prognostic models for ER-negative breast cancer patients. This combined clinical and genomics model approach can also be used to build predictors of therapy responsiveness, and could ultimately be applied to other tumor types.
Figures



References
-
- van't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AAM, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT. et al.Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

