Epigenetics in alternative pre-mRNA splicing
- PMID: 21215366
- PMCID: PMC3038581
- DOI: 10.1016/j.cell.2010.11.056
Epigenetics in alternative pre-mRNA splicing
Abstract
Alternative splicing plays critical roles in differentiation, development, and disease and is a major source for protein diversity in higher eukaryotes. Analysis of alternative splicing regulation has traditionally focused on RNA sequence elements and their associated splicing factors, but recent provocative studies point to a key function of chromatin structure and histone modifications in alternative splicing regulation. These insights suggest that epigenetic regulation determines not only what parts of the genome are expressed but also how they are spliced.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures

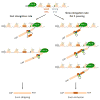



References
-
- Aebi M, Weissmann C. Precision and orderliness in splicing. Trends Genet. 1987;3:102–107.
-
- Allo M, Buggiano V, Fededa JP, Petrillo E, Schor I, de la Mata M, Agirre E, Plass M, Eyras E, Abou Elela S, Klinck R, Chabot B, Kornblihtt AR. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat Struct Mol Biol. 2009;16:717–724. - PubMed
-
- Attanasio C, David A, Neerman-Arbez M. Outcome of donor splice site mutations accounting for congenital afibrinogenemia reflects order of intron removal in the fibrinogen alpha gene (FGA) Blood. 2003;101:1851–1856. - PubMed
-
- Auboeuf D, Honig A, Berget SM, O’Malley BW. Coordinate regulation of transcription and splicing by steroid receptor coregulators. Science. 2002;298:416–419. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

