Structural biology of retroviral DNA integration
- PMID: 21216426
- PMCID: PMC3640404
- DOI: 10.1016/j.virol.2010.12.008
Structural biology of retroviral DNA integration
Abstract
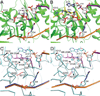
Three-dimensional macromolecular structures shed critical light on biological mechanism and facilitate development of small molecule inhibitors. Clinical success of raltegravir, a potent inhibitor of HIV-1 integrase, demonstrated the utility of this viral DNA recombinase as an antiviral target. A variety of partial integrase structures reported in the past 16 years have been instrumental and very informative to the field. Nonetheless, because integrase protein fragments are unable to functionally engage the viral DNA substrate critical for strand transfer inhibitor binding, the early structures did little to materially impact drug development efforts. However, recent results based on prototype foamy virus integrase have fully reversed this trend, as a number of X-ray crystal structures of active integrase-DNA complexes revealed key mechanistic details and moreover established the foundation of HIV-1 integrase strand transfer inhibitor action. In this review we discuss the landmarks in the progress of integrase structural biology during the past 17 years.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures

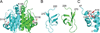
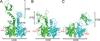
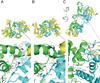

References
-
- Bao KK, Wang H, Miller JK, Erie DA, Skalka AM, Wong I. Functional oligomeric state of avian sarcoma virus integrase. J. Biol. Chem. 2003;278:1323–1327. - PubMed
-
- Baumann H, Knapp S, Lundbäck T, Ladenstein R, Härd T. Solution structure and DNA-binding properties of a thermostable protein from the archaeon Sulfolobus solfataricus. Nat. Struct. Biol. 1994;1:808–819. - PubMed
-
- Baus J, Liu L, Heggestad AD, Sanz S, Fletcher BS. Hyperactive transposase mutants of the Sleeping Beauty transposon. Mol. Ther. 2005;12:1148–1156. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

