New insights into the signaling mechanism of the pH-responsive, membrane-integrated transcriptional activator CadC of Escherichia coli
- PMID: 21216950
- PMCID: PMC3060519
- DOI: 10.1074/jbc.M110.196923
New insights into the signaling mechanism of the pH-responsive, membrane-integrated transcriptional activator CadC of Escherichia coli
Abstract
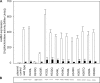
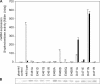
The membrane-integrated transcriptional regulator CadC of Escherichia coli activates expression of the cadBA operon at low external pH with concomitantly available lysine, providing adaptation to mild acidic stress. CadC is a representative of the ToxR-like proteins that combine sensory, signal transduction, and DNA-binding activities within a single polypeptide. Although several ToxR-like regulators such as CadC, as well as the main regulator of Vibrio cholerae virulence, ToxR itself, which activate gene expression at acidic pH, have been intensively investigated, their molecular activation mechanism is still unclear. In this study, a structure-guided mutational analysis was performed to elucidate the mechanism by which CadC detects acidification of the external milieu. Thus, a cluster of negatively charged amino acids (Asp-198, Asp-200, Glu-461, Glu-468, and Asp-471) was found to be crucial for pH detection. These amino acids form a negatively charged patch on the surface of the periplasmic domain of CadC that stretches across its two subdomains. The results of different combinations of amino acid replacements within this patch indicated that the N-terminal subdomain integrates and transduces the signals coming from both subdomains to the transmembrane domain. Alterations in the phospholipid composition did not influence pH-dependent cadBA expression, and therefore, interplay of the acidic surface patch with the negatively charged headgroups is unlikely. Models are discussed according to which protonation of these acidic amino acid side chains reduces repulsive forces between the two subdomains and/or between two monomers within a CadC dimer and thereby enables receptor activation upon lowering of the environmental pH.
Figures


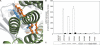



References
-
- Bearson S., Bearson B., Foster J. W. (1997) FEMS Microbiol. Lett. 147, 173–180 - PubMed
-
- Auger E. A., Redding K. E., Plumb T., Childs L. C., Meng S. Y., Bennett G. N. (1989) Mol. Microbiol. 3, 609–620 - PubMed
-
- Soksawatmaekhin W., Kuraishi A., Sakata K., Kashiwagi K., Igarashi K. (2004) Mol. Microbiol. 51, 1401–1412 - PubMed
-
- Fritz G., Koller C., Burdack K., Tetsch L., Haneburger I., Jung K., Gerland U. (2009) J. Mol. Biol. 393, 272–286 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

