HSP70 natively and specifically associates with an N-terminal dermcidin-derived peptide that contains an HLA-A*03 antigenic epitope
- PMID: 21216960
- PMCID: PMC3069480
- DOI: 10.1074/jbc.M110.179630
HSP70 natively and specifically associates with an N-terminal dermcidin-derived peptide that contains an HLA-A*03 antigenic epitope
Abstract
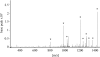
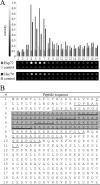
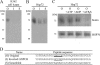
Tumor cells very often have elevated expression of HSP70, the anti-apoptotic properties of which contribute to overall tumor survival. Independent of its anti-apoptotic properties, HSP70 was also suggested to be involved in the antigen presentation process by chaperoning cytosolic peptides, thus protecting them from rapid degradation and securing the peptide pool for further processing. In this study, we identified a 33-amino acid N-terminal dermcidin (DCD)-derived peptide from the repertoire of in vivo HSP70-associated peptides isolated from a leukemic cell line, K562. The DCD peptide has been previously shown to be involved in tumorigenesis, to increase tumor survival rate, to improve tumor stress resistance, and to aid growth. We show that HSP70 is a specific binding partner for the DCD prosurvival peptide and define an ATP-dependent DCD-binding site (GNPCH). We also identify an HLA-A*03 antigenic epitope within the DCD peptide, which follows and partially overlaps the HSP70-binding site (CHEASAAQK). This study describes the interaction between HSP70 and the DCD-derived prosurvival peptide, an interaction that may direct the peptide toward antigen presentation and independently contribute to the prosurvival mechanism mediated by DCD.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

