A fast, lock-free approach for efficient parallel counting of occurrences of k-mers
- PMID: 21217122
- PMCID: PMC3051319
- DOI: 10.1093/bioinformatics/btr011
A fast, lock-free approach for efficient parallel counting of occurrences of k-mers
Abstract
Motivation: Counting the number of occurrences of every k-mer (substring of length k) in a long string is a central subproblem in many applications, including genome assembly, error correction of sequencing reads, fast multiple sequence alignment and repeat detection. Recently, the deep sequence coverage generated by next-generation sequencing technologies has caused the amount of sequence to be processed during a genome project to grow rapidly, and has rendered current k-mer counting tools too slow and memory intensive. At the same time, large multicore computers have become commonplace in research facilities allowing for a new parallel computational paradigm.
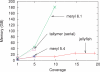
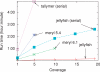
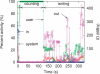
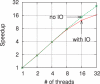
Results: We propose a new k-mer counting algorithm and associated implementation, called Jellyfish, which is fast and memory efficient. It is based on a multithreaded, lock-free hash table optimized for counting k-mers up to 31 bases in length. Due to their flexibility, suffix arrays have been the data structure of choice for solving many string problems. For the task of k-mer counting, important in many biological applications, Jellyfish offers a much faster and more memory-efficient solution.
Availability: The Jellyfish software is written in C++ and is GPL licensed. It is available for download at http://www.cbcb.umd.edu/software/jellyfish.
Figures
References
-
- Campagna D, et al. RAP: a new computer program for de novo identification of repeated sequences in whole genomes. Bioinformatics. 2005;21:582–588. - PubMed
-
- Cormen T, et al. Introduction to Algorithms. Cambridge, MA: MIT Press; 1990. Chapter 12.
-
- Dean J, Ghemawat S. MapReduce: simplified data processing on large clusters. Commun. ACM. 2008;51:107–113.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

