Molecular mechanism of Ena/VASP-mediated actin-filament elongation
- PMID: 21217643
- PMCID: PMC3034019
- DOI: 10.1038/emboj.2010.348
Molecular mechanism of Ena/VASP-mediated actin-filament elongation
Abstract
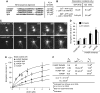
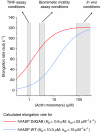
Ena/VASP proteins are implicated in a variety of fundamental cellular processes including axon guidance and cell migration. In vitro, they enhance elongation of actin filaments, but at rates differing in nearly an order of magnitude according to species, raising questions about the molecular determinants of rate control. Chimeras from fast and slow elongating VASP proteins were generated and their ability to promote actin polymerization and to bind G-actin was assessed. By in vitro TIRF microscopy as well as thermodynamic and kinetic analyses, we show that the velocity of VASP-mediated filament elongation depends on G-actin recruitment by the WASP homology 2 motif. Comparison of the experimentally observed elongation rates with a quantitative mathematical model moreover revealed that Ena/VASP-mediated filament elongation displays a saturation dependence on the actin monomer concentration, implying that Ena/VASP proteins, independent of species, are fully saturated with actin in vivo and generally act as potent filament elongators. Moreover, our data showed that spontaneous addition of monomers does not occur during processive VASP-mediated filament elongation on surfaces, suggesting that most filament formation in cells is actively controlled.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


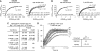


References
-
- Bachmann C, Fischer L, Walter U, Reinhard M (1999) The EVH2 domain of the vasodilator-stimulated phosphoprotein mediates tetramerization, F-actin binding, and actin bundle formation. J Biol Chem 274: 23549–23557 - PubMed
-
- Bear JE, Loureiro JJ, Libova I, Fässler R, Wehland J, Gertler FB (2000) Negative regulation of fibroblast motility by Ena/VASP proteins. Cell 101: 717–728 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

