Discovery and preclinical validation of salivary transcriptomic and proteomic biomarkers for the non-invasive detection of breast cancer
- PMID: 21217834
- PMCID: PMC3013113
- DOI: 10.1371/journal.pone.0015573
Discovery and preclinical validation of salivary transcriptomic and proteomic biomarkers for the non-invasive detection of breast cancer
Abstract
Background: A sensitive assay to identify biomarkers using non-invasively collected clinical specimens is ideal for breast cancer detection. While there are other studies showing disease biomarkers in saliva for breast cancer, our study tests the hypothesis that there are breast cancer discriminatory biomarkers in saliva using de novo discovery and validation approaches. This is the first study of this kind and no other study has engaged a de novo biomarker discovery approach in saliva for breast cancer detection. In this study, a case-control discovery and independent preclinical validations were conducted to evaluate the performance and translational utilities of salivary transcriptomic and proteomic biomarkers for breast cancer detection.
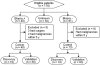
Methodology/principal findings: Salivary transcriptomes and proteomes of 10 breast cancer patients and 10 matched controls were profiled using Affymetrix HG-U133-Plus-2.0 Array and two-dimensional difference gel electrophoresis (2D-DIGE), respectively. Preclinical validations were performed to evaluate the discovered biomarkers in an independent sample cohort of 30 breast cancer patients and 63 controls using RT-qPCR (transcriptomic biomarkers) and quantitative protein immunoblot (proteomic biomarkers). Transcriptomic and proteomic profiling revealed significant variations in salivary molecular biomarkers between breast cancer patients and matched controls. Eight mRNA biomarkers and one protein biomarker, which were not affected by the confounding factors, were pre-validated, yielding an accuracy of 92% (83% sensitive, 97% specific) on the preclinical validation sample set.
Conclusions: Our findings support that transcriptomic and proteomic signatures in saliva can serve as biomarkers for the non-invasive detection of breast cancer. The salivary biomarkers possess discriminatory power for the detection of breast cancer, with high specificity and sensitivity, which paves the way for prediction model validation study followed by pivotal clinical validation.
Conflict of interest statement
Figures

References
-
- Hinestrosa MC, Dickersin K, Klein P, Mayer M, Noss K, et al. Shaping the future of biomarker research in breast cancer to ensure clinical relevance. Nat Rev Cancer. 2007;7:309–315. - PubMed
-
- Levenson VV. Biomarkers for early detection of breast cancer: what, when, and where? Biochim Biophys Acta. 2007;1770:847–856. - PubMed
-
- Etzioni R, Urban N, Ramsey S, McIntosh M, Schwartz S, et al. The case for early detection. Nat Rev Cancer. 2003;3:243–252. - PubMed
-
- Cheang MC, van de Rijn M, Nielsen TO. Gene expression profiling of breast cancer. Annu Rev Pathol. 2008;3:67–97. - PubMed
-
- Morris SR, Carey LA. Gene expression profiling in breast cancer. Curr Opin Oncol. 2007;19:547–551. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

