Structural basis for the nucleotide-dependent dimerization of the large G protein atlastin-1/SPG3A
- PMID: 21220294
- PMCID: PMC3038741
- DOI: 10.1073/pnas.1012792108
Structural basis for the nucleotide-dependent dimerization of the large G protein atlastin-1/SPG3A
Abstract
The large GTPase atlastin belongs to the dynamin superfamily that has been widely implicated in facilitating membrane tubulation, fission, and in select cases, fusion. Mutations spread across atlastin isoform 1 (atlastin-1) have been identified in patients suffering from hereditary spastic paraplegia (HSP), a neurodegenerative disorder affecting motor neuron function in the lower extremities. On a molecular level, atlastin-1 associates with high membrane curvature and fusion events at the endoplasmic reticulum and cis-Golgi. Here we report crystal structures of atlastin-1 comprising the G and middle domains in two different conformations. Although the orientation of the middle domain relative to the G domain is different in the two structures, both reveal dimeric assemblies with a common, GDP-bound G domain dimer. In contrast, dimer formation in solution is observed only in the presence of GTP and transition state analogs, similar to other G proteins that are activated by nucleotide-dependent dimerization. Analyses of solution scattering data suggest that upon nucleotide binding, the protein adopts a somewhat extended, dimeric conformation that is reminiscent of one of the two crystal structures. These structural studies suggest a model for nucleotide-dependent regulation of atlastin with implications for membrane fusion. This mechanism is affected in several mutants associated with HSP, providing insights into disease pathogenesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
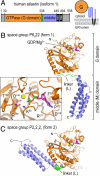
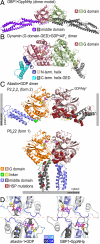
 is shown (PDB ID code 2X2E) (4). (C) Atlastin-1. Dimers observed in crystal lattices with P212121 symmetry (form 2, Upper) and with P6522 symmetry (form 1, Lower) are shown. GDP and Mg2+ positions are indicated and are shown as sticks and spheres, respectively. Hypothetical membrane positions based on the location of the C terminus of the cytosolic domain are indicated. Positions of missense mutation associated with HSP are shown as residues displayed as red spheres in both forms. (D) Nucleotide arrangement and G domain dimer interface. The G domain dimer interaces of atlastin-1 (form 1; Left) and GBP1 (Right; PDB ID code 2BC9) (3) are shown. The protomers are colored in light and dark gray, with the nucleotide-binding regions (G1–G4) shown in shades of blue. Nucleotide and Mg2+ are shown as sticks and spheres, respectively. The dashed, red line separates the dimer halves, marking the interfacial region.
is shown (PDB ID code 2X2E) (4). (C) Atlastin-1. Dimers observed in crystal lattices with P212121 symmetry (form 2, Upper) and with P6522 symmetry (form 1, Lower) are shown. GDP and Mg2+ positions are indicated and are shown as sticks and spheres, respectively. Hypothetical membrane positions based on the location of the C terminus of the cytosolic domain are indicated. Positions of missense mutation associated with HSP are shown as residues displayed as red spheres in both forms. (D) Nucleotide arrangement and G domain dimer interface. The G domain dimer interaces of atlastin-1 (form 1; Left) and GBP1 (Right; PDB ID code 2BC9) (3) are shown. The protomers are colored in light and dark gray, with the nucleotide-binding regions (G1–G4) shown in shades of blue. Nucleotide and Mg2+ are shown as sticks and spheres, respectively. The dashed, red line separates the dimer halves, marking the interfacial region.
Comment in
-
Structural insights into membrane fusion at the endoplasmic reticulum.Proc Natl Acad Sci U S A. 2011 Feb 8;108(6):2175-6. doi: 10.1073/pnas.1019194108. Epub 2011 Jan 28. Proc Natl Acad Sci U S A. 2011. PMID: 21278333 Free PMC article. No abstract available.
References
-
- Praefcke GJK, McMahon HT. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat Rev Mol Cell Biol. 2004;5:133–147. - PubMed
-
- Gasper R, Meyer S, Gotthardt K, Sirajuddin M, Wittinghofer A. It takes two to tango: Regulation of G proteins by dimerization. Nat Rev Mol Cell Biol. 2009;10:423–429. - PubMed
-
- Prakash B, Praefcke GJ, Renault L, Wittinghofer A, Herrmann C. Structure of human guanylate-binding protein 1 representing a unique class of GTP-binding proteins. Nature. 2000;403:567–571. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

