Masking the 5' terminal nucleotides of the hepatitis C virus genome by an unconventional microRNA-target RNA complex
- PMID: 21220300
- PMCID: PMC3044371
- DOI: 10.1073/pnas.1012464108
Masking the 5' terminal nucleotides of the hepatitis C virus genome by an unconventional microRNA-target RNA complex
Abstract
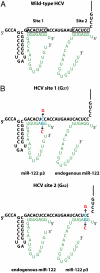
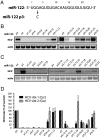
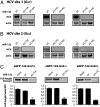
Hepatitis C virus subverts liver-specific microRNA, miR-122, to upregulate viral RNA abundance in both infected cultured cells and in the liver of infected chimpanzees. These findings have identified miR-122 as an attractive antiviral target. Thus, it is imperative to know whether a distinct functional complex exists between miR-122 and the viral RNA versus its normal cellular target mRNAs. Toward this goal, effects on viral RNA abundance of mutated miR-122 duplex molecules, bound at each of the two target sites in the viral genome, were compared to effects on microRNA- or siRNA-mediated regulation of reporter target mRNAs. It was found that miR-122 formed an unusual microRNA complex with the viral RNA that is distinct from miR-122 complexes with reporter mRNAs. Notably, miR-122 forms an oligomeric complex in which one miR-122 molecule binds to the 5' terminus of the hepatitis C virus (HCV) RNA with 3' overhanging nucleotides, masking the 5' terminal sequences of the HCV genome. Furthermore, specific internal nucleotides as well as the 3' terminal nucleotides in miR-122 were absolutely required for maintaining HCV RNA abundance but not for microRNA function. Both miR-122 molecules utilize similar internal nucleotides to interact with the viral genome, creating a bulge and tail in the miR-122 molecules, revealing tandemly oriented oligomeric RNA complexes. These findings suggest that miR-122 protects the 5' terminal viral sequences from nucleolytic degradation or from inducing innate immune responses to the RNA terminus. Finally, this remarkable microRNA-mRNA complex could be targeted with compounds that inactivate miR-122 or interfere with this unique RNA structure.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Comment in
-
End game: getting the most out of microRNAs.Proc Natl Acad Sci U S A. 2011 Feb 22;108(8):3101-2. doi: 10.1073/pnas.1019613108. Epub 2011 Feb 9. Proc Natl Acad Sci U S A. 2011. PMID: 21307312 Free PMC article. No abstract available.
References
-
- Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5:453–463. - PubMed
-
- Hoofnagle JH. Course and outcome of hepatitis C. Hepatology. 2002;36:S21–S29. - PubMed
-
- Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

