A quantitative assay for measuring mRNA decapping by splinted ligation reverse transcription polymerase chain reaction: qSL-RT-PCR
- PMID: 21220548
- PMCID: PMC3039152
- DOI: 10.1261/rna.2436411
A quantitative assay for measuring mRNA decapping by splinted ligation reverse transcription polymerase chain reaction: qSL-RT-PCR
Abstract
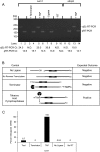
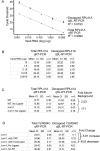
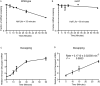
The degradation of messenger RNA is a critical node of gene regulation. A major pathway of mRNA decay is initiated by shortening of the poly(A) tail, followed by removal of the 5' cap structure (decapping) and subsequent degradation. Decapping is an important determinate in the destruction of many transcripts. Detailed kinetic analysis of in vivo decapping rates is necessary to understand how this step is regulated. Importantly, the product of decapping is recalcitrant for investigation, in part due to its transient nature. As such, little in vivo kinetic information is available. Here we report the development of an assay that measures decapping of mRNAs by combining splinted ligation and quantitative RT-PCR (qSL-RT-PCR). We apply this method to determine the decapping rate constant for a natural mRNA in vivo for the first time. The qSL-RT-PCR assay may be adapted for use on any mRNA, providing a new tool to study regulation of mRNA decay.
Figures

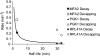
References
-
- Beelman CA, Stevens A, Caponigro G, LaGrandeur TE, Hatfield L, Fortner DM, Parker R 1996. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature 382: 642–646 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
