Ecologically and evolutionarily important SNPs identified in natural populations
- PMID: 21220761
- PMCID: PMC3098511
- DOI: 10.1093/molbev/msr004
Ecologically and evolutionarily important SNPs identified in natural populations
Abstract
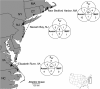
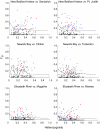
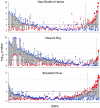
Evolution by natural selection acts on natural populations amidst migration, gene-by-environmental interactions, constraints, and tradeoffs, which affect the rate and frequency of adaptive change. We asked how many and how rapidly loci change in populations subject to severe, recent environmental changes. To address these questions, we used genomic approaches to identify randomly selected single nucleotide polymorphisms (SNPs) with evolutionarily significant patterns in three natural populations of Fundulus heteroclitus that inhabit and have adapted to highly polluted Superfund sites. Three statistical tests identified 1.4-2.5% of SNPs that were significantly different from the neutral model in each polluted population. These nonneutral patterns in populations adapted to highly polluted environments suggest that these loci or closely linked loci are evolving by natural selection. One SNP identified in all polluted populations using all tests is in the gene for the xenobiotic metabolizing enzyme, cytochrome P4501A (CYP1A), which has been identified previously as being refractory to induction in the three highly polluted populations. Extrapolating across the genome, these data suggest that rapid evolutionary change in natural populations can involve hundreds of loci, a few of which will be shared in independent events.
Figures

References
-
- Adams SM, Lindmeier JB, Duvernell DD. Microsatellite analysis of the phylogeography, Pleistocene history and secondary contact hypotheses for the killifish, Fundulus heteroclitus. Mol Ecol. 2006;15:1109–1123. - PubMed
-
- Beaumont M, Nichols R. Evaluating loci for use in the genetic analysis of population structure. Proc R Soc Lond Biol Sci. 1996;263:1619–1626.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

