Controlled translocation of individual DNA molecules through protein nanopores with engineered molecular brakes
- PMID: 21222450
- PMCID: PMC3391008
- DOI: 10.1021/nl1038874
Controlled translocation of individual DNA molecules through protein nanopores with engineered molecular brakes
Abstract
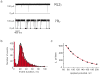
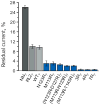
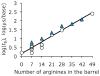
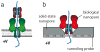
Protein nanopores may provide a cheap and fast technology to sequence individual DNA molecules. However, the electrophoretic translocation of ssDNA molecules through protein nanopores has been too rapid for base identification. Here, we show that the translocation of DNA molecules through the α-hemolysin protein nanopore can be slowed controllably by introducing positive charges into the lumen of the pore by site directed mutagenesis. Although the residual ionic current during DNA translocation is insufficient for direct base identification, we propose that the engineered pores might be used to slow down DNA in hybrid systems, for example, in combination with solid-state nanopores.
Figures

References
-
- Drmanac R, Sparks AB, Callow MJ, Halpern AL, Burns NL, Kermani BG, Carnevali P, Nazarenko I, Nilsen GB, Yeung G, Dahl F, Fernandez A, Staker B, Pant KP, Baccash J, Borcherding AP, Brownley A, Cedeno R, Chen L, Chernikoff D, Cheung A, Chirita R, Curson B, Ebert JC, Hacker CR, Hartlage R, Hauser B, Huang S, Jiang Y, Karpinchyk V, Koenig M, Kong C, Landers T, Le C, Liu J, McBride CE, Morenzoni M, Morey RE, Mutch K, Perazich H, Perry K, Peters BA, Peterson J, Pethiyagoda CL, Pothuraju K, Richter C, Rosenbaum AM, Roy S, Shafto J, Sharanhovich U, Shannon KW, Sheppy CG, Sun M, Thakuria JV, Tran A, Vu D, Zaranek AW, Wu X, Drmanac S, Oliphant AR, Banyai WC, Martin B, Ballinger DG, Church GM, Reid CA. Science. 2010;327(5961):78–81. - PubMed
-
- Bonetta L. Cell. 2010;141(6):917–9. - PubMed
-
- Branton D, Deamer DW, Marziali A, Bayley H, Benner SA, Butler T, Di Ventra M, Garaj S, Hibbs A, Huang X, Jovanovich SB, Krstic PS, Lindsay S, Ling XS, Mastrangelo CH, Meller A, Oliver JS, Pershin YV, Ramsey JM, Riehn R, Soni GV, Tabard-Cossa V, Wanunu M, Wiggin M, Schloss JA. Nature Biotechnology. 2008;26:1146–1153. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

