Bioinformatic evidence for a widely distributed, ribosomally produced electron carrier precursor, its maturation proteins, and its nicotinoprotein redox partners
- PMID: 21223593
- PMCID: PMC3023750
- DOI: 10.1186/1471-2164-12-21
Bioinformatic evidence for a widely distributed, ribosomally produced electron carrier precursor, its maturation proteins, and its nicotinoprotein redox partners
Abstract
Background: Enzymes in the radical SAM (rSAM) domain family serve in a wide variety of biological processes, including RNA modification, enzyme activation, bacteriocin core peptide maturation, and cofactor biosynthesis. Evolutionary pressures and relationships to other cellular constituents impose recognizable grammars on each class of rSAM-containing system, shaping patterns in results obtained through various comparative genomics analyses.
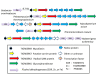
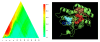
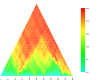
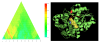
Results: An uncharacterized gene cluster found in many Actinobacteria and sporadically in Firmicutes, Chloroflexi, Deltaproteobacteria, and one Archaeal plasmid contains a PqqE-like rSAM protein family that includes Rv0693 from Mycobacterium tuberculosis. Members occur clustered with a strikingly well-conserved small polypeptide we designate "mycofactocin," similar in size to bacteriocins and PqqA, precursor of pyrroloquinoline quinone (PQQ). Partial Phylogenetic Profiling (PPP) based on the distribution of these markers identifies the mycofactocin cluster, but also a second tier of high-scoring proteins. This tier, strikingly, is filled with up to thirty-one members per genome from three variant subfamilies that occur, one each, in three unrelated classes of nicotinoproteins. The pattern suggests these variant enzymes require not only NAD(P), but also the novel gene cluster. Further study was conducted using SIMBAL, a PPP-like tool, to search these nicotinoproteins for subsequences best correlated across multiple genomes to the presence of mycofactocin. For both the short chain dehydrogenase/reductase (SDR) and iron-containing dehydrogenase families, aligning SIMBAL's top-scoring sequences to homologous solved crystal structures shows signals centered over NAD(P)-binding sites rather than over substrate-binding or active site residues. Previous studies on some of these proteins have revealed a non-exchangeable NAD cofactor, such that enzymatic activity in vitro requires an artificial electron acceptor such as N,N-dimethyl-4-nitrosoaniline (NDMA) for the enzyme to cycle.
Conclusions: Taken together, these findings suggest that the mycofactocin precursor is modified by the Rv0693 family rSAM protein and other enzymes in its cluster. It becomes an electron carrier molecule that serves in vivo as NDMA and other artificial electron acceptors do in vitro. Subclasses from three different nicotinoprotein families show "only-if" relationships to mycofactocin because they require its presence. This framework suggests a segregated redox pool in which mycofactocin mediates communication among enzymes with non-exchangeable cofactors.
Figures

References
-
- Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crecy-Lagard V, Diaz N, Disz T, Edwards R. et al.The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33(17):5691–5702. doi: 10.1093/nar/gki866. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

