Non-invasive prenatal assessment of trisomy 21 by multiplexed maternal plasma DNA sequencing: large scale validity study
- PMID: 21224326
- PMCID: PMC3019239
- DOI: 10.1136/bmj.c7401
Non-invasive prenatal assessment of trisomy 21 by multiplexed maternal plasma DNA sequencing: large scale validity study
Abstract
Objectives: To validate the clinical efficacy and practical feasibility of massively parallel maternal plasma DNA sequencing to screen for fetal trisomy 21 among high risk pregnancies clinically indicated for amniocentesis or chorionic villus sampling.
Design: Diagnostic accuracy validated against full karyotyping, using prospectively collected or archived maternal plasma samples.
Setting: Prenatal diagnostic units in Hong Kong, United Kingdom, and the Netherlands.
Participants: 753 pregnant women at high risk for fetal trisomy 21 who underwent definitive diagnosis by full karyotyping, of whom 86 had a fetus with trisomy 21. Intervention Multiplexed massively parallel sequencing of DNA molecules in maternal plasma according to two protocols with different levels of sample throughput: 2-plex and 8-plex sequencing.
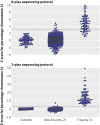
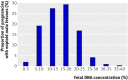
Main outcome measures: Proportion of DNA molecules that originated from chromosome 21. A trisomy 21 fetus was diagnosed when the z score for the proportion of chromosome 21 DNA molecules was >3. Diagnostic sensitivity, specificity, positive predictive value, and negative predictive value were calculated for trisomy 21 detection.
Results: Results were available from 753 pregnancies with the 8-plex sequencing protocol and from 314 pregnancies with the 2-plex protocol. The performance of the 2-plex protocol was superior to that of the 8-plex protocol. With the 2-plex protocol, trisomy 21 fetuses were detected at 100% sensitivity and 97.9% specificity, which resulted in a positive predictive value of 96.6% and negative predictive value of 100%. The 8-plex protocol detected 79.1% of the trisomy 21 fetuses and 98.9% specificity, giving a positive predictive value of 91.9% and negative predictive value of 96.9%.
Conclusion: Multiplexed maternal plasma DNA sequencing analysis could be used to rule out fetal trisomy 21 among high risk pregnancies. If referrals for amniocentesis or chorionic villus sampling were based on the sequencing test results, about 98% of the invasive diagnostic procedures could be avoided.
Conflict of interest statement
Competing interests: All authors have completed the Unified Competing Interest form at
Figures

References
-
- Driscoll DA, Gross S. Clinical practice. Prenatal screening for aneuploidy. N Engl J Med 2009;360:2556-62. - PubMed
-
- Tabor A, Philip J, Madsen M, Bang J, Obel EB, Norgaard-Pedersen B. Randomised controlled trial of genetic amniocentesis in 4606 low-risk women. Lancet 1986;1:1287-93. - PubMed
-
- Snijders RJ, Noble P, Sebire N, Souka A, Nicolaides KH. UK multicentre project on assessment of risk of trisomy 21 by maternal age and fetal nuchal-translucency thickness at 10-14 weeks of gestation. Fetal Medicine Foundation First Trimester Screening Group. Lancet 1998;352:343-6. - PubMed
-
- Kagan KO, Wright D, Baker A, Sahota D, Nicolaides KH. Screening for trisomy 21 by maternal age, fetal nuchal translucency thickness, free beta-human chorionic gonadotropin and pregnancy-associated plasma protein-A. Ultrasound Obstet Gynecol 2008;31:618-24. - PubMed
-
- Malone FD, Canick JA, Ball RH, Nyberg DA, Comstock CH, Bukowski R, et al. First- trimester or second-trimester screening, or both, for Down’s syndrome. N Engl J Med 2005;353:2001-11. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
