Neural stem and progenitor cells shorten S-phase on commitment to neuron production
- PMID: 21224845
- PMCID: PMC3105305
- DOI: 10.1038/ncomms1155
Neural stem and progenitor cells shorten S-phase on commitment to neuron production
Abstract
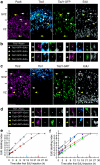
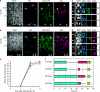
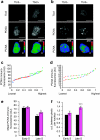
During mammalian cerebral cortex development, the G1-phase of the cell cycle is known to lengthen, but it has been unclear which neural stem and progenitor cells are affected. In this paper, we develop a novel approach to determine cell-cycle parameters in specific classes of neural stem and progenitor cells, identified by molecular markers rather than location. We found that G1 lengthening was associated with the transition from stem cell-like apical progenitors to fate-restricted basal (intermediate) progenitors. Unexpectedly, expanding apical and basal progenitors exhibit a substantially longer S-phase than apical and basal progenitors committed to neuron production. Comparative genome-wide gene expression analysis of expanding versus committed progenitor cells revealed changes in key factors of cell-cycle regulation, DNA replication and repair and chromatin remodelling. Our findings suggest that expanding neural stem and progenitor cells invest more time during S-phase into quality control of replicated DNA than those committed to neuron production.
Figures



References
-
- Kriegstein A. R. & Götz M. Radial glia diversity: a matter of cell fate. Glia 43, 37–43 (2003). - PubMed
-
- Götz M. & Huttner W. B. The cell biology of neurogenesis. Nat. Rev. Mol. Cell Biol. 6, 777–788 (2005). - PubMed
-
- Farkas L. M. & Huttner W. B. The cell biology of neural stem and progenitor cells and its significance for their proliferation versus differentiation during mammalian brain development. Curr. Opin. Cell Biol. 20, 707–715 (2008). - PubMed
-
- Noctor S. C., Martinez-Cerdeno V., Ivic L. & Kriegstein A. R. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat. Neurosci. 7, 136–144 (2004). - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

