Temporal change in genetic integrity suggests loss of local adaptation in a wild Atlantic salmon (Salmo salar) population following introgression by farmed escapees
- PMID: 21224876
- PMCID: PMC3131974
- DOI: 10.1038/hdy.2010.165
Temporal change in genetic integrity suggests loss of local adaptation in a wild Atlantic salmon (Salmo salar) population following introgression by farmed escapees
Abstract
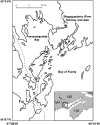
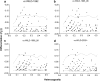
In some wild Atlantic salmon populations, rapid declines in numbers of wild returning adults has been associated with an increase in the prevalence of farmed salmon. Studies of phenotypic variation have shown that interbreeding between farmed and wild salmon may lead to loss of local adaptation. Yet, few studies have attempted to assess the impact of interbreeding at the genome level, especially among North American populations. Here, we document temporal changes in the genetic makeup of the severely threatened Magaguadavic River salmon population (Bay of Fundy, Canada), a population that might have been impacted by interbreeding with farmed salmon for nearly 20 years. Wild and farmed individuals caught entering the river from 1980 to 2005 were genotyped at 112 single-nucleotide polymorphisms (SNPs), and/or eight microsatellite loci, to scan for potential shifts in adaptive genetic variation. No significant temporal change in microsatellite-based estimates of allele richness or gene diversity was detected in the wild population, despite its precipitous decline in numbers over the last two decades. This might reflect the effect of introgression from farmed salmon, which was corroborated by temporal change in linkage-disequilibrium. Moreover, SNP genome scans identified a temporal decrease in candidate loci potentially under directional selection. Of particular interest was a SNP previously shown to be strongly associated with an important quantitative trait locus for parr mark number, which retained its genetic distinctiveness between farmed and wild fish longer than other outliers. Overall, these results indicate that farmed escapees have introgressed with wild Magaguadavic salmon resulting in significant alteration of the genetic integrity of the native population, including possible loss of adaptation to wild conditions.
Figures


References
-
- Agapow PM, Burt A. Indices of multilocus linkage disequilibrium. Mol Ecol Notes. 2001;1:101–102.
-
- Allendorf FW, Luikart G. Conservation and the Genetics of Populations. Blackwell Publishing: Oxford, UK; 2007.
-
- Beaumont MA, Nichols RA. Evaluating loci for use in the genetic analysis of population structure. Proc R Soc Lond B Biol Sci. 1996;263:1619–1626.
-
- Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. Laboratoire Génome, Population, Interactions: CNRS UMR 5000. Université de Montpellier II: Montpellier, France; 2001. GENETIX 4.02, logiciel sous windows TM pour la génétique des populations.
-
- Boulding EG, Culling M, Glebe B, Berg PR, Lien S, Moen T. Conservation genomics of Atlantic salmon: SNPs associated with QTLs for adaptive traits in parr from four trans-Atlantic backcrosses. Heredity. 2008;101:381–391. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

