Dhx34 and Nbas function in the NMD pathway and are required for embryonic development in zebrafish
- PMID: 21227923
- PMCID: PMC3089463
- DOI: 10.1093/nar/gkq1319
Dhx34 and Nbas function in the NMD pathway and are required for embryonic development in zebrafish
Abstract
The nonsense-mediated mRNA decay (NMD) pathway is a highly conserved surveillance mechanism that is present in all eukaryotes. It prevents the synthesis of truncated proteins by selectively degrading mRNAs harbouring premature termination codons (PTCs). The core NMD effectors were originally identified in genetic screens in Saccharomyces cerevisae and in the nematode Caenorhabditis elegans, and subsequently by homology searches in other metazoans. A genome-wide RNAi screen in C. elegans resulted in the identification of two novel NMD genes that are essential for proper embryonic development. Their human orthologues, DHX34 and NAG/NBAS, are required for NMD in human cells. Here, we find that the zebrafish genome encodes orthologues of DHX34 and NAG/NBAS. We show that the morpholino-induced depletion of zebrafish Dhx34 and Nbas proteins results in severe developmental defects and reduced embryonic viability. We also found that Dhx34 and Nbas are required for degradation of PTC-containing mRNAs in zebrafish embryos. The phenotypes observed in both Dhx34 and Nbas morphants are similar to defects in Upf1, Smg-5- or Smg-6- depleted embryos, suggesting that these factors affect the same pathway and confirming that zebrafish embryogenesis requires an active NMD pathway.
© The Author(s) 2011. Published by Oxford University Press.
Figures
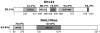



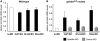
References
-
- Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 2007;76:51–74. - PubMed
-
- Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet. 2004;36:1073–1078. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

