Identification of MST1/STK4 and SULF1 proteins as autoantibody targets for the diagnosis of colorectal cancer by using phage microarrays
- PMID: 21228115
- PMCID: PMC3047148
- DOI: 10.1074/mcp.M110.001784
Identification of MST1/STK4 and SULF1 proteins as autoantibody targets for the diagnosis of colorectal cancer by using phage microarrays
Abstract
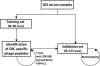
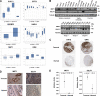
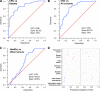
The characterization of the humoral response in cancer patients is becoming a practical alternative to improve early detection. We prepared phage microarrays containing colorectal cancer cDNA libraries to identify phage-expressed peptides recognized by tumor-specific autoantibodies from patient sera. From a total of 1536 printed phages, 128 gave statistically significant values to discriminate cancer patients from control samples. From this, 43 peptide sequences were unique following DNA sequencing. Six phages containing homologous sequences to STK4/MST1, SULF1, NHSL1, SREBF2, GRN, and GTF2I were selected to build up a predictor panel. A previous study with high-density protein microarrays had identified STK4/MST1 as a candidate biomarker. An independent collection of 153 serum samples (50 colorectal cancer sera and 103 reference samples, including healthy donors and sera from other related pathologies) was used as a validation set to study prediction capability. A combination of four phages and two recombinant proteins, corresponding to MST1 and SULF1, achieved an area under the curve of 0.86 to correctly discriminate cancer from healthy sera. Inclusion of sera from other different neoplasias did not change significantly this value. For early stages (A+B), the corrected area under the curve was 0.786. Moreover, we have demonstrated that MST1 and SULF1 proteins, homologous to phage-peptide sequences, can replace the original phages in the predictor panel, improving their diagnostic accuracy.
Figures

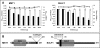
References
-
- Edwards B. K., Ward E., Kohler B. A., Eheman C., Zauber A. G., Anderson R. N., Jemal A., Schymura M. J., Lansdorp-Vogelaar I., Seeff L. C., van Ballegooijen M., Goede S. L., Ries L. A. (2010) Annual report to the nation on the status of cancer, 1975–2006, featuring colorectal cancer trends and impact of interventions (risk factors, screening, and treatment) to reduce future rates. Cancer 116, 544–573 - PMC - PubMed
-
- Winawer S. J. (2007) The multidisciplinary management of gastrointestinal cancer. Colorectal cancer screening. Best Pract. Res. Clin. Gastroenterol. 21, 1031–1048 - PubMed
-
- Duffy M. J., van Dalen A., Haglund C., Hansson L., Holinski-Feder E., Klapdor R., Lamerz R., Peltomaki P., Sturgeon C., Topolcan O. (2007) Tumour markers in colorectal cancer: European Group on Tumour Markers (EGTM) guidelines for clinical use. Eur. J. Cancer 43, 1348–1360 - PubMed
-
- Barderas R., Babel I., Casal J. I. (2010) Colorectal cancer proteomics, molecular characterization and biomarker discovery. Proteomics Clin. Appl. 4, 159–178 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

